Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene - PubMed (original) (raw)
. 2007 Aug 22;129(33):10220-8.
doi: 10.1021/ja072185g. Epub 2007 Aug 2.
Affiliations
- PMID: 17672459
- PMCID: PMC2566970
- DOI: 10.1021/ja072185g
Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene
Kexiao Guo et al. J Am Chem Soc. 2007.
Abstract
A polypurine (guanine)/polypyrimidine (cytosine)-rich sequence within the proximal promoter region of the human RET oncogene has been shown to be essential for RET basal transcription. Specifically, the G-rich strand within this region consists of five consecutive runs of guanines, which is consistent with the general motif capable of forming intramolecular G-quadruplexes. Here we demonstrate that, in the presence of 100 mM K+, this G-rich strand has the ability to adopt two intramolecular G-quadruplex structures in vitro. Moreover, comparative circular dichroism (CD) and DMS footprinting studies have revealed that the 3'-G-quadruplex structure is a parallel-type intramolecular structure containing three G-tetrads. The G-quadruplex-interactive agents TMPyP4 and telomestatin further stabilize this G-quadruplex structure. In addition, we demonstrate that the complementary C-rich strand forms an i-motif structure in vitro, as shown by CD spectroscopy and chemical footprinting. This 19-mer duplex sequence is predicted to form stable intramolecular G-quadruplex and i-motif species having minimum symmetrical loop sizes of 1:3:1 and 2:3:2, respectively. Together, our results indicate that stable G-quadruplex and i-motif structures can form within the proximal promoter region of the human RET oncogene, suggesting that these secondary structures play an important role in transcriptional regulation of this gene.
Figures
Figure 1
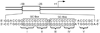
Schematic diagram of the proximal promoter region of the RET proto-oncogene. Two GC boxes are labeled. Five runs of guanines (I, II, III, IV and V) are underlined.
Figure 2
Schematic diagram of a G-quadruplex and an i-motif structure. (A) Four guanines form a G-tetrad through Hoogsteen bonds, and three G-tetrads form a parallel G-quadruplex structure in the c-Myc promoter. (B) Hemiprotonated cytosine–cytosine base pair. Two C+–C pairs form an intermolecular i-motif structure.
Figure 3
Taq polymerase stop assay of the G-rich region of the RET promoter. (A) The sequences of the RET-Pol1 and RET-Pol2 templates used in the Taq polymerase stop assay. The five runs of guanines are labeled I–V. The full-length product, minor stop site, major stop site, and primer site are indicated with arrows. (B) and (C) show the Taq polymerase stop assay with RET-Pol1 and RET-Pol2, respectively, with increasing concentrations of KCl (lane 1, 0 mM KCl; lane 2, 25 mM KCl; lane 3, 50 mM KCl, lane 4, 100 mM KCl). For both (B) and (C), the full-length product, minor stop site, major stop site, and primer site are indicated with arrows on the right. The sequencing reaction for C is shown on the left side of each gel.
Figure 4
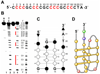
Taq polymerase stop assay with the wild-type template (RET-Pol2) and mutant templates. The wild-type template (RET-Pol2) is shown on the top, with arrows indicating the primer site, stop site and full-length product. The four consecutive runs of guanines are underlined in bold, with guanines 3, 8, 13, and 18 indicated. For mutant templates (A3, A8, A13, A18, T3, T8, T13, and T18), the letter refers to the mutated nucleotide and the number refers to the position (3, 8, 13, or 18) on RET-Pol2. For example, A3 means a G-to-A mutation at position 3 on RET-Pol2. The Taq polymerase stop assay was carried out in the absence of (−) or in the presence of 100 mM KCl (+). Sequencing reactions for G and C are shown on the left side of the gel. Lanes 1 and 2 are reactions with the wild-type template (RET-Pol2). Lanes 3–18 are reactions with the different mutant templates. The full-length product, stop site, and primer site are indicated with arrows on the right.
Figure 5
DMS footprinting, CD spectra, and melting curve of the G-quadruplex structure formed by RET31. (A) DMS footprinting of an intramolecular G-quadruplex structure. Lanes 1 and 2 are the AG and TC sequencing reactions, lane 3 is the DMS footprinting in the absence of KCl, and lane 4 is the one in the presence of 100 mM KCl. The partial sequence of RET31 is shown to the right of the gel. The protected guanines are indicated by open circles. Guanines 1, 9, 11, 19, and 20 show enhanced cleavage (closed circles) and partial protection (semi-open circles). (B) The superimposition of the CD spectrum of RET31 and that of myc-1245. (C) The CD melting curve of the G-quadruplex structure. (D) The proposed G-quadruplex structure formed by RET31
Figure 6
DNA polymerase stop assay using RET-Pol2 as the template with the addition of increasing concentrations of TMPyP4 (0, 1, 2 and 5 µM) and telomestatin (0, 0.1, 0.5 and 2 µM) in 10 mM KCl/NaCl. Sequencing reactions for G and C are shown on the left side of the gel. (B) CD spectra of RET31 with increasing concentrations of TMPyP4 from 1-mole equivalence to 4-mole equivalence in Tris-HCl buffer (20 mM, pH 7.6), 10 mM KCl. (C) CD spectra of RET31 with increasing concentrations of telomestatin from 1-mole equivalence to 4-mole equivalence in Tris-HCl buffer (20 mM, pH 7.6), 10 mM KCl.
Figure 7
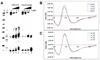
CD spectra of RET1 recorded at room temperature in Tris-acetate buffer (50 mM). (A) Fifteen spectra were collected, and selected spectra at pH 4.35, pH 5.50, pH 6.00, pH 6.50, pH 7.10, and pH 8.00 are shown. (B) pH dependence of the molar ellipticity at 288 nm (C) The CD melting curves of RET1 at three different pHs.
Figure 8
Br2 footprinting of the unimolecular i-motif structures formed by RET1. (A) Sequence of RET1 oligonucleotide used in Br2 protection experiments. (B) Autoradiogram of 20% denaturing PAGE experiment to determine cytosine residues involved in base pairing and intercalation to form intramolecular i-motif structures. Lanes 1 (AG) and 2 (TC) represent the purine- and pyrimidine-specific reactions, respectively, to generate sequencing markers. Lanes 3–5 represent reactions without Br2, with Br2 at pH 5.2, and with Br2 at pH 8.0, respectively. (C) Summary of Br2 cleavage. The open circles represent the protected cytosine residues, while closed circles represent enhanced cleavage at the cytosine residues. (D) Folding pattern of the proposed i-motif structure formed by RET1.
Figure 9
Comparison of truncated G-quadruplex-forming sequences within selected gene promoters.
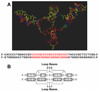
Figure 10
(A) Stable low energy model of the RET promoter sequence (−66 to −19) with i-motif, G-quadruplex, and duplex DNA regions, displayed as capped sticks (adenine = green, guanine = red, thymine = blue, and cytosine = yellow) with potassium ions as CPK model (white). Hydrogen atoms are not shown for clarity. The corresponding RET promoter sequence is shown below the model. DNA bases involved in i-motif and G-quadruplex structure formation are shown in red. (B) Symmetrical arrangement of RET C-rich and G-rich sequences, indicating loop bases and bases participating in the formation of i-motifs and G-quadruplexes (boxes).
Similar articles
- Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops.
Benabou S, Ferreira R, Aviñó A, González C, Lyonnais S, Solà M, Eritja R, Jaumot J, Gargallo R. Benabou S, et al. Biochim Biophys Acta. 2014 Jan;1840(1):41-52. doi: 10.1016/j.bbagen.2013.08.028. Epub 2013 Sep 6. Biochim Biophys Acta. 2014. PMID: 24012973 - The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents.
Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. Sun D, et al. Mol Cancer Ther. 2008 Apr;7(4):880-9. doi: 10.1158/1535-7163.MCT-07-2119. Mol Cancer Ther. 2008. PMID: 18413801 Free PMC article. - Structure of the biologically relevant G-quadruplex in the c-MYC promoter.
Yang D, Hurley LH. Yang D, et al. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):951-68. doi: 10.1080/15257770600809913. Nucleosides Nucleotides Nucleic Acids. 2006. PMID: 16901825 Review. - Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions.
Qin Y, Hurley LH. Qin Y, et al. Biochimie. 2008 Aug;90(8):1149-71. doi: 10.1016/j.biochi.2008.02.020. Epub 2008 Feb 29. Biochimie. 2008. PMID: 18355457 Free PMC article. Review.
Cited by
- G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA.
Dhakal S, Yu Z, Konik R, Cui Y, Koirala D, Mao H. Dhakal S, et al. Biophys J. 2012 Jun 6;102(11):2575-84. doi: 10.1016/j.bpj.2012.04.024. Epub 2012 Jun 5. Biophys J. 2012. PMID: 22713573 Free PMC article. - Structural insights into G-quadruplexes: towards new anticancer drugs.
Yang D, Okamoto K. Yang D, et al. Future Med Chem. 2010 Apr;2(4):619-46. doi: 10.4155/fmc.09.172. Future Med Chem. 2010. PMID: 20563318 Free PMC article. Review. - Systematic investigation of sequence requirements for DNA i-motif formation.
Školáková P, Renčiuk D, Palacký J, Krafčík D, Dvořáková Z, Kejnovská I, Bednářová K, Vorlíčková M. Školáková P, et al. Nucleic Acids Res. 2019 Mar 18;47(5):2177-2189. doi: 10.1093/nar/gkz046. Nucleic Acids Res. 2019. PMID: 30715498 Free PMC article. - Recognition and discrimination of DNA quadruplexes by acridine-peptide conjugates.
Redman JE, Granadino-Roldán JM, Schouten JA, Ladame S, Reszka AP, Neidle S, Balasubramanian S. Redman JE, et al. Org Biomol Chem. 2009 Jan 7;7(1):76-84. doi: 10.1039/b814682a. Epub 2008 Oct 30. Org Biomol Chem. 2009. PMID: 19081949 Free PMC article. - Stability and kinetics of G-quadruplex structures.
Lane AN, Chaires JB, Gray RD, Trent JO. Lane AN, et al. Nucleic Acids Res. 2008 Oct;36(17):5482-515. doi: 10.1093/nar/gkn517. Epub 2008 Aug 21. Nucleic Acids Res. 2008. PMID: 18718931 Free PMC article. Review.
References
- Takahashi M, Buma Y, Iwamoto T, Inaguma Y, Ikeda H, Hiai H. Oncogene. 1988;3:571–578. - PubMed
- Jhiang SM. Oncogene. 2000;19:5590–5597. - PubMed
- Arighi E, Borrello MG, Sariola H. Cytokine Growth Factor Rev. 2005;16:441–467. - PubMed
- Kouvaraki MA, Shapiro SE, Perrier ND, Cote GJ, Gagel RF, Hoff AO, Sherman SI, Lee JE, Evans DB. Thyroid. 2005;15:531–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA95060/CA/NCI NIH HHS/United States
- P50 CA095060-070004/CA/NCI NIH HHS/United States
- CA94166/CA/NCI NIH HHS/United States
- P50 CA095060/CA/NCI NIH HHS/United States
- R01 CA094166-05/CA/NCI NIH HHS/United States
- R01 CA094166/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous