Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup - PubMed (original) (raw)
Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup
Nadin Rohland et al. PLoS Biol. 2007 Aug.
Abstract
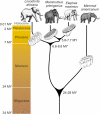
We have sequenced the complete mitochondrial genome of the extinct American mastodon (Mammut americanum) from an Alaskan fossil that is between 50,000 and 130,000 y old, extending the age range of genomic analyses by almost a complete glacial cycle. The sequence we obtained is substantially different from previously reported partial mastodon mitochondrial DNA sequences. By comparing those partial sequences to other proboscidean sequences, we conclude that we have obtained the first sequence of mastodon DNA ever reported. Using the sequence of the mastodon, which diverged 24-28 million years ago (mya) from the Elephantidae lineage, as an outgroup, we infer that the ancestors of African elephants diverged from the lineage leading to mammoths and Asian elephants approximately 7.6 mya and that mammoths and Asian elephants diverged approximately 6.7 mya. We also conclude that the nuclear genomes of the African savannah and forest elephants diverged approximately 4.0 mya, supporting the view that these two groups represent different species. Finally, we found the mitochondrial mutation rate of proboscideans to be roughly half of the rate in primates during at least the last 24 million years.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
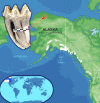
Figure 1. Appearance and Location of Origin of the Mastodon Tooth (IK-99–237) Used for Sequencing of the mtDNA Genome
The scale bar shows centimeters.
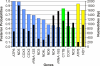
Figure 2. Ratio of the Number of Transitions to the Number of Transversions
The six points correspond to the ratio between species of Elephantidae (first two points), between Elephantidae and Mammut, between Proboscidea and (Dugong, Procavia), between Uranotheria and Bos, and between Mammalia and Struthio. The corresponding divergence times for proboscideans are given in Table 2. Dugong and hyrax diverged from Proboscidea 65 mya [5,9]. We used 105 mya for the Uranotheria–ruminant split ([9] and citations therein). The mammalian–bird divergence was set to 313 mya (the mean value of the confidence interval of [62]; but see also [63,65]). The arrows indicate the points of interest. The uncertainty of the divergence times is not taken into account because the goal was to show the general trend.
Figure 3. Gene-by-Gene Topology
Colored bars indicate the posterior probabilities for each respective topology. Blue indicates a Mammuthus–Elephas sister group relationship, green indicates a Mammuthus–Loxodonta sister group relationship, and yellow indicates an Elephas–Loxodonta sister group relationship. Black bars show the length of the respective genes in base pairs.
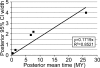
Figure 4. Data Saturation as Determined by the 95% CI Width as a Function of Posterior Mean Time
The high correlation coefficient of this plot suggests that the sequence data are highly informative. However, it cannot be excluded that the accuracy of divergence time estimates could be further improved by additional sequence data. The graph corresponds to the whole genome analysis. The partitioned data give a slightly higher correlation coefficient of 0.89 (not shown).
Figure 5. Phylogeny and Divergence Times Determined by This Study for the Four Proboscidean Species Analyzed
Similar articles
- Phylogenetic resolution within the Elephantidae using fossil DNA sequence from the American mastodon (Mammut americanum) as an outgroup.
Yang H, Golenberg EM, Shoshani J. Yang H, et al. Proc Natl Acad Sci U S A. 1996 Feb 6;93(3):1190-4. doi: 10.1073/pnas.93.3.1190. Proc Natl Acad Sci U S A. 1996. PMID: 8577738 Free PMC article. - Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants.
Rohland N, Reich D, Mallick S, Meyer M, Green RE, Georgiadis NJ, Roca AL, Hofreiter M. Rohland N, et al. PLoS Biol. 2010 Dec 21;8(12):e1000564. doi: 10.1371/journal.pbio.1000564. PLoS Biol. 2010. PMID: 21203580 Free PMC article. - The mastodon mitochondrial genome: a mammoth accomplishment.
Roca AL. Roca AL. Trends Genet. 2008 Feb;24(2):49-52. doi: 10.1016/j.tig.2007.11.005. Epub 2008 Jan 14. Trends Genet. 2008. PMID: 18192067 Review. - Forest elephant mitochondrial genomes reveal that elephantid diversification in Africa tracked climate transitions.
Brandt AL, Ishida Y, Georgiadis NJ, Roca AL. Brandt AL, et al. Mol Ecol. 2012 Mar;21(5):1175-89. doi: 10.1111/j.1365-294X.2012.05461.x. Epub 2012 Jan 19. Mol Ecol. 2012. PMID: 22260276 - Elephant natural history: a genomic perspective.
Roca AL, Ishida Y, Brandt AL, Benjamin NR, Zhao K, Georgiadis NJ. Roca AL, et al. Annu Rev Anim Biosci. 2015;3:139-67. doi: 10.1146/annurev-animal-022114-110838. Epub 2014 Dec 8. Annu Rev Anim Biosci. 2015. PMID: 25493538 Review.
Cited by
- Rapid evolution of genes with anti-cancer functions during the origins of large bodies and cancer resistance in elephants.
Bowman J, Lynch VJ. Bowman J, et al. bioRxiv [Preprint]. 2024 Feb 29:2024.02.27.582135. doi: 10.1101/2024.02.27.582135. bioRxiv. 2024. PMID: 38463968 Free PMC article. Preprint. - A genetic glimpse of the Chinese straight-tusked elephants.
Lin H, Hu J, Baleka S, Yuan J, Chen X, Xiao B, Song S, Du Z, Lai X, Hofreiter M, Sheng G. Lin H, et al. Biol Lett. 2023 Jul;19(7):20230078. doi: 10.1098/rsbl.2023.0078. Epub 2023 Jul 19. Biol Lett. 2023. PMID: 37463654 Free PMC article. - Genetic features of Sri Lankan elephant, Elephas maximus maximus Linnaeus revealed by high throughput sequencing of mitogenome and ddRAD-seq.
Sooriyabandara MGC, Jayasundara JMSM, Marasinghe MSLRP, Hathurusinghe HABM, Bandaranayake AU, Jayawardane KANC, Nilanthi RMR, Rajapakse RC, Bandaranayake PCG. Sooriyabandara MGC, et al. PLoS One. 2023 Jun 13;18(6):e0285572. doi: 10.1371/journal.pone.0285572. eCollection 2023. PLoS One. 2023. PMID: 37310948 Free PMC article. - Comparison of the gut microbiome and resistome in captive African and Asian elephants on the same diet.
Feng X, Hua R, Zhang W, Liu Y, Luo C, Li T, Chen X, Zhu H, Wang Y, Lu Y. Feng X, et al. Front Vet Sci. 2023 Feb 16;10:986382. doi: 10.3389/fvets.2023.986382. eCollection 2023. Front Vet Sci. 2023. PMID: 36875997 Free PMC article. - Aging: What We Can Learn From Elephants.
Chusyd DE, Ackermans NL, Austad SN, Hof PR, Mielke MM, Sherwood CC, Allison DB. Chusyd DE, et al. Front Aging. 2021 Aug 26;2:726714. doi: 10.3389/fragi.2021.726714. eCollection 2021. Front Aging. 2021. PMID: 35822016 Free PMC article. Review.
References
- Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, et al. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 2001;409:704–707. - PubMed
- Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. - PubMed
- Shoshani J. Understanding proboscidean evolution: A formidable task. Trends Ecol Evol. 1998;13:480–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources