Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport - PubMed (original) (raw)
Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport
Nafiseh Sabri et al. J Cell Biol. 2007.
Abstract
The phenylanine-glycine (FG)-rich regions of several nucleoporins both bind to nuclear transport receptors and collectively provide a diffusion barrier to the nuclear pores. However, the in vivo roles of FG nucleoporins in transport remain unclear. We have inactivated 30 putative nucleoporins in cultured Drosophila melanogaster S2 cells by RNA interference and analyzed the phenotypes on importin alpha/beta-mediated import and CRM1-dependent protein export. The fly homologues of FG nucleoporins Nup358, Nup153, and Nup54 are selectively required for import. The FG repeats of Nup153 are necessary for its function in transport, whereas the remainder of the protein maintains pore integrity. Inactivation of the CRM1 cofactor RanBP3 decreased the nuclear accumulation of CRM1 and protein export. We report a surprisingly antagonistic relationship between RanBP3 and the Nup214 FG region in determining CRM1 localization and its function in protein export. Our data suggest that peripheral metazoan FG nucleoporins have distinct functions in nuclear protein transport events.
Figures
Figure 1.
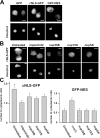
cNLS-GFP import defects in Nup358, Nup153, and Nup54 RNAi cells. (A) Localization of GFP, cNLS-GFP, and GFP-NES in S2 cells. Hoechst staining visualizes nuclei. (B) Cells expressing cNLS-GFP were treated with importin β, Nup358, Nup153, or Nup54 dsRNAs. (C) Ratios of nuclear to cytoplasmic cNLS-GFP (left) and GFP-NES (right) intensities in untreated and RNAi cells. dsRNA treatments reduced the nuclear accumulation of cNLS-GFP compared with untreated cells (P < 0.0001 by pair-wise _t_ test). GFP-NES distribution was only affected in c_rmi_ cells (P > 0.05 by pair-wise t test for the nucleoporins). Error bars indicate SD. 30–35 cells were quantified for each treatment. Bars, 5 μm.
Figure 2.
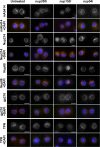
Localization of nuclear pore components in cells defective in cNLS-GFP import. Cells subjected to Nup358, Nup153, or Nup54 dsRNA treatment stained with mAb414 and antibodies against Nup214, Nup88, gp210, and TPR. DAPI staining visualizes nuclei. Bars, 5 μm.
Figure 3.
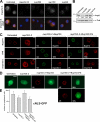
Separable roles of Nup153 in pore integrity and cNLS import. (A) Cells treated with importin β, Nup358, Nup153, or Nup54 dsRNAs stained for importin β. DAPI staining visualizes nuclei. (B) Western blot of extracts from Nup358, Nup54, and Nup153 RNAi cells probed with anti–importin β and antitubulin antibodies. Importin β intensities were normalized against tubulin. Numbers indicate the relative levels of importin β in each sample. (C) Function of full-length Nup153 and Nup153ΔFG in pore integrity and importin β localization. The first column shows untreated cells. The second column shows cells treated with dsRNA against the 3′ untranslated region of Nup153 (nup153i-2). The other columns show cells treated in parallel and transfected with either V5-Nup153 or V5-Nup153ΔFG plasmids. Cells were stained for V5, Nup214, TPR, and importin β. V5 staining visualizes the expression of Nup153 fusion proteins. All panels show confocal sections. (D) cNLS-GFP cells treated as in C stained for V5 (red). GFP flourescence is shown in green. cNLS-GFP distribution was restored in only 10% of cells expressing Nup153ΔFG. (E) Quantification of nuclear to cytoplasmic cNLS-GFP intensity ratios in Nup153 RNAi cells. The expression of V5-Nup153 (P < 0.0001 by pair-wise _t_ test) but not of V5-Nup153ΔFG (P > 0.05 by pair-wise t test) restored cNLS-GFP distribution. Error bars indicate SD. 20–25 cells were quantified in each case. Bars (A), 5 μm; (C and D) 2.5 μm.
Figure 4.
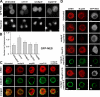
Antagonistic functions of RanBP3 and Nup214 in GFP-NES export and CRM1 localization. (A) GFP-NES–expressing cells were treated with CRM1, RanBP3, or Nup214 dsRNAs. Hoechst staining visualizes nuclei. (B) Ratios of nuclear to cytoplasmic GFP-NES in untreated cells and cells subjected to CRM1, RanBP3, Nup214, or Nup88 dsRNA treatment. CRM1 and RanBP3 RNAi cells showed the increased nuclear accumulation of GFP-NES compared with untreated cells (P < 0.0001 by pair-wise _t_ test). Nup214 and Nup88 RNAi did not significantly alter GFP-NES distribution (P > 0.05 by pair-wise t test). Error bars indicate SD. 30–35 cells were quantified for each treatment. (C) Cells treated with RanBP3 dsRNA, Nup88 dsRNA, Nup214 dsRNA, or a combination of RanBP3 and Nup214 dsRNAs stained for CRM1 (red) and mAb414 (green). (D) Cells treated with RanBP3 dsRNA or dsRNAs against both RanBP3 and the 3′ untranslated region of Nup214 (ranbp3i + nup214i-2) were transfected either with V5-Nup214 or V5-Nup214ΔFG plasmids. Cells were stained for V5, Nup88, and CRM1. The right column shows GFP-NES cells treated as in other columns stained for V5 and analyzed for GFP localization. V5 staining visualizes the expression of Nup214 fusion protein. In all cases, V5 labeling is presented in green. Bars (A), 5 μm; (C and D) 2.5 μm.
Figure 5.
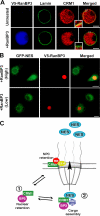
RanBP3 overexpression attracts CRM1 from the NPC. (A) Cells expressing V5-RanBP3 stained for V5 (blue), lamin (green), and CRM1 (red). Insets show magnified images of the boxed areas. (B) GFP-NES–expressing cells transfected with the V5-RanBP3 plasmid stained for V5 (red) and analyzed for the localization of GFP (green). (C) Model illustrating the postulated dynamic equilibrium between two interchangeable pools of CRM1. One is anchored by Nup214 at the NPC, and the other is retained in the nucleus by RanBP3. The model proposes two functions of RanBP3. (1) It retains CRM1 inside the nucleus. This function is antagonized by the NPC-anchoring activity of Nup214. (2) RanBP3 also promotes the assembly of NES cargo complexes and export. Bars (A), 2.5 μm; (B) 5 μm.
Similar articles
- Nup214 is required for CRM1-dependent nuclear protein export in vivo.
Hutten S, Kehlenbach RH. Hutten S, et al. Mol Cell Biol. 2006 Sep;26(18):6772-85. doi: 10.1128/MCB.00342-06. Mol Cell Biol. 2006. PMID: 16943420 Free PMC article. - The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila.
Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. Xylourgidis N, et al. J Cell Sci. 2006 Nov 1;119(Pt 21):4409-19. doi: 10.1242/jcs.03201. Epub 2006 Oct 10. J Cell Sci. 2006. PMID: 17032737 - The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport.
Wälde S, Kehlenbach RH. Wälde S, et al. Trends Cell Biol. 2010 Aug;20(8):461-9. doi: 10.1016/j.tcb.2010.05.001. Epub 2010 Jun 4. Trends Cell Biol. 2010. PMID: 20627572 Review. - Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex.
Patel SS, Belmont BJ, Sante JM, Rexach MF. Patel SS, et al. Cell. 2007 Apr 6;129(1):83-96. doi: 10.1016/j.cell.2007.01.044. Cell. 2007. PMID: 17418788 - Converging on the function of intrinsically disordered nucleoporins in the nuclear pore complex.
Peleg O, Lim RY. Peleg O, et al. Biol Chem. 2010 Jul;391(7):719-30. doi: 10.1515/BC.2010.092. Biol Chem. 2010. PMID: 20482319 Review.
Cited by
- Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila.
Panda D, Pascual-Garcia P, Dunagin M, Tudor M, Hopkins KC, Xu J, Gold B, Raj A, Capelson M, Cherry S. Panda D, et al. Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):E3890-9. doi: 10.1073/pnas.1410087111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197089 Free PMC article. - Nucleoporin 54 contributes to homologous recombination repair and post-replicative DNA integrity.
Rodriguez-Berriguete G, Granata G, Puliyadi R, Tiwana G, Prevo R, Wilson RS, Yu S, Buffa F, Humphrey TC, McKenna WG, Higgins GS. Rodriguez-Berriguete G, et al. Nucleic Acids Res. 2018 Sep 6;46(15):7731-7746. doi: 10.1093/nar/gky569. Nucleic Acids Res. 2018. PMID: 29986057 Free PMC article. - The Nup153-Nup50 protein interface and its role in nuclear import.
Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. Makise M, et al. J Biol Chem. 2012 Nov 9;287(46):38515-22. doi: 10.1074/jbc.M112.378893. Epub 2012 Sep 24. J Biol Chem. 2012. PMID: 23007389 Free PMC article. - Misregulation of Nucleoporins 98 and 96 leads to defects in protein synthesis that promote hallmarks of tumorigenesis.
Pulianmackal AJ, Kanakousaki K, Flegel K, Grushko OG, Gourley E, Rozich E, Buttitta LA. Pulianmackal AJ, et al. Dis Model Mech. 2022 Mar 1;15(3):dmm049234. doi: 10.1242/dmm.049234. Epub 2022 Mar 16. Dis Model Mech. 2022. PMID: 35107131 Free PMC article. - COPI vesicle transport is a common requirement for tube expansion in Drosophila.
Jayaram SA, Senti KA, Tiklová K, Tsarouhas V, Hemphälä J, Samakovlis C. Jayaram SA, et al. PLoS One. 2008 Apr 9;3(4):e1964. doi: 10.1371/journal.pone.0001964. PLoS One. 2008. PMID: 18398480 Free PMC article.
References
- Bayliss, R., T. Littlewood, L.A. Strawn, S.R. Wente, and M. Stewart. 2002. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277:50597–50606. - PubMed
- Bernad, R., D. Engelsma, H. Sanderson, H. Pickersgill, and M. Fornerod. 2006. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J. Biol. Chem. 281:19378–19386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous