RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? - PubMed (original) (raw)
Comment
RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage?
Laura A Lindsey-Boltz et al. Proc Natl Acad Sci U S A. 2007.
No abstract available
Conflict of interest statement
The authors declare no conflict of interest.
Figures
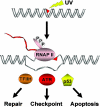
Fig. 1.
RNAP II as the universal high-specificity damage sensor for three major cellular responses to bulky DNA lesions, such as the cyclobutane pyrimidine dimers induced by UV light. RNAP II arrests at a dimer site in the transcribed strand. The resulting structure recruits proteins that initiate repair, cell cycle checkpoints, or apoptosis.
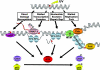
Fig. 2.
Potential DNA damage detection mechanisms for initiating checkpoint responses. The primary damage itself may be recognized by damage-specific proteins that may activate the checkpoint directly. Alternatively, the stalled RNAP II constitutes the checkpoint signal and recruits checkpoint kinase ATR. RPA-coated single-stranded DNA gaps generated by nucleotide excision repair and the extensive RPA-coated single-stranded DNA generated by replication forks stalled at a damage site are known to be strong signals for checkpoint activation.
Comment on
- RPA and ATR link transcriptional stress to p53.
Derheimer FA, O'Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. Derheimer FA, et al. Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12778-83. doi: 10.1073/pnas.0705317104. Epub 2007 Jul 6. Proc Natl Acad Sci U S A. 2007. PMID: 17616578 Free PMC article.
Similar articles
- DNA photodamage recognition by RNA polymerase II.
Brueckner F, Cramer P. Brueckner F, et al. FEBS Lett. 2007 Jun 19;581(15):2757-60. doi: 10.1016/j.febslet.2007.05.014. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17521634 Review. - Damage control: DNA repair, transcription, and the ubiquitin-proteasome system.
Daulny A, Tansey WP. Daulny A, et al. DNA Repair (Amst). 2009 Apr 5;8(4):444-8. doi: 10.1016/j.dnarep.2009.01.017. Epub 2009 Mar 9. DNA Repair (Amst). 2009. PMID: 19272841 Review. - Blockage of RNA polymerase II at a cyclobutane pyrimidine dimer and 6-4 photoproduct.
Mei Kwei JS, Kuraoka I, Horibata K, Ubukata M, Kobatake E, Iwai S, Handa H, Tanaka K. Mei Kwei JS, et al. Biochem Biophys Res Commun. 2004 Aug 6;320(4):1133-8. doi: 10.1016/j.bbrc.2004.06.066. Biochem Biophys Res Commun. 2004. PMID: 15249207 - Shedding UV light on alternative splicing.
Marengo MS, Garcia-Blanco MA. Marengo MS, et al. Cell. 2009 May 15;137(4):600-2. doi: 10.1016/j.cell.2009.04.054. Cell. 2009. PMID: 19450507 - CPD damage recognition by transcribing RNA polymerase II.
Brueckner F, Hennecke U, Carell T, Cramer P. Brueckner F, et al. Science. 2007 Feb 9;315(5813):859-62. doi: 10.1126/science.1135400. Science. 2007. PMID: 17290000
Cited by
- A Nobel-Winning Scientist: Aziz Sancar and the Impact of his Work on the Molecular Pathology of Neoplastic Diseases.
Pehlivanoglu B, Aysal A, Kececi SD, Ekmekci S, Erdogdu IH, Ertunc O, Gundogdu B, Talu CK, Sahin Y, Toper MH. Pehlivanoglu B, et al. Turk Patoloji Derg. 2021;37(2):93-105. doi: 10.5146/tjpath.2020.01504. Turk Patoloji Derg. 2021. PMID: 33973640 Free PMC article. Review. - Multiple roles of CTDK-I throughout the cell.
Srivastava R, Duan R, Ahn SH. Srivastava R, et al. Cell Mol Life Sci. 2019 Jul;76(14):2789-2797. doi: 10.1007/s00018-019-03118-0. Epub 2019 Apr 29. Cell Mol Life Sci. 2019. PMID: 31037337 Free PMC article. Review. - Mechanism of Rad26-assisted rescue of stalled RNA polymerase II in transcription-coupled repair.
Yan C, Dodd T, Yu J, Leung B, Xu J, Oh J, Wang D, Ivanov I. Yan C, et al. Nat Commun. 2021 Dec 1;12(1):7001. doi: 10.1038/s41467-021-27295-4. Nat Commun. 2021. PMID: 34853308 Free PMC article. - Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome.
Li W, Adebali O, Yang Y, Selby CP, Sancar A. Li W, et al. Proc Natl Acad Sci U S A. 2018 Apr 10;115(15):E3408-E3415. doi: 10.1073/pnas.1801687115. Epub 2018 Mar 26. Proc Natl Acad Sci U S A. 2018. PMID: 29581276 Free PMC article. - RNA polymerase between lesion bypass and DNA repair.
Deaconescu AM. Deaconescu AM. Cell Mol Life Sci. 2013 Dec;70(23):4495-509. doi: 10.1007/s00018-013-1384-3. Epub 2013 Jun 27. Cell Mol Life Sci. 2013. PMID: 23807206 Free PMC article. Review.
References
- Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. - PubMed
- Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources