The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor - PubMed (original) (raw)
The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor
Birger Herzog et al. Mol Endocrinol. 2007 Nov.
Abstract
The liver X receptors (LXRs) are nuclear receptors that play important roles in the regulation of lipid metabolism. In this study, we demonstrate that receptor-interacting protein 140 (RIP140) is a cofactor for LXR in liver. Analysis of RIP140 null mice and hepatocytes depleted of RIP140 indicate that the cofactor is essential for the ability of LXR to activate the expression of a set of genes required for lipogenesis. Furthermore we demonstrate that RIP140 is required for the ability of LXR to repress the expression of the phosphoenolpyruvate carboxykinase gene in Fao cells and mice. Thus, we conclude that the function of RIP140 as a cofactor for LXR in liver varies according to the target genes and metabolic process, serving as a coactivator in lipogenesis but as a corepressor in gluconeogenesis.
Figures
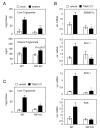
Fig. 1. Hepatic lipid metabolism in WT and RIP140 null mice
A, Triglyceride amounts in liver and plasma of WT and RIP140 null mice maintained on chow or western diet for 4 weeks. Results are expressed as the mean ± SEM of 5-10 animals in each group. ***p<0.001 versus WT fed with western diet. B, WT and RIP140 null mice were maintained on a standard chow diet and fed a synthetic LXR agonist (T0901317) or vehicle alone for 3 days (50 mg/kg/day). Gene expression was determined by real-time PCR analysis. Shown are relative mRNA levels normalized to cyclophilin as mean ± SEM of 4-5 animals in each group. C, Hepatic triglyceride content in WT and RIP140 null mice maintained on a standard chow diet and fed T0901317 or vehicle alone. *p<0.05, **p<0.01versus WT treated with T0901317.
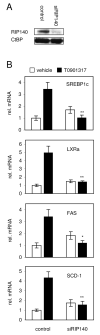
Fig. 2. Function of RIP140 in primary hepatocytes
A, Total lipid content was measured in primary hepatocytes isolated from wild-type (WT) or RIP140 null mice (RIP KO). Cells were stimulated for 24 h with T0901317 or vehicle alone. Data is expressed as fold change compared to wild-type control samples. B, Cells were treated as described above and gene expression was determined by real-time PCR. Shown are relative mRNA levels normalized to cyclophilin as mean ± SEM, *p<0.05 versus WT treated with T0901317.
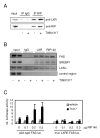
Fig. 3. RIP140 function in LXR-mediated lipogenesis
A, HuH7 cells were treated with siRNA for RIP140 (siRIP140) or control (pAd-GFP) as indicated. After 48 h RIP140 and CtBP protein was detected by Western blotting. B, Gene expression of lipogenic genes was determined following treatment with 1μM T0901317 or vehicle alone for 12-16 h in HuH7 cells depleted of RIP140 (siRIP140) or controls (pAd-GFP). mRNA levels were measured by real-time PCR and normalized to cyclophilin. Data are expressed as fold change relative to control samples and represents the average of 3 experiments performed in triplicate ± SEM. *p<0.05, **p<0.01 versus control treated with T0901317.
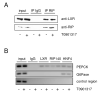
Fig. 4. Interaction of LXR and RIP140 in HuH7 cells
A, Coimmunopreciptiation assays were performed with nuclear extracts from HuH7 cells treated with 1μM T0901317 or vehicle alone for 2 h. B, Chromatin immunopreciptiation assay for LXR and RIP140 in HuH7 cells treated with T0901317 or vehicle alone for 2 h. Purified DNA from precipitated chromatin fragments were amplified by PCR using primers encompassing the LXREs in the promoter region of the indicated genes or a distal region of the FAS gene. The distal region of the FAS gene does not harbour a LXRE and was used as a control. C, Cotransfection of WT or mutant FAS reporter genes, LXRα and increasing amounts of RIP140 expression plasmids. Cells were treated with T0901317 as indicated. Data represent the mean luciferase activity of 3-4 experiments ± SEM.
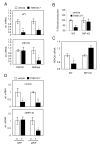
Fig. 5. RIP140 function in the regulation of gluconeogenesis
A, PEPCK and G6Pase gene expression in WT and RIP140 null mice treated with T0901317 (50 mg/kg/day) or vehicle alone for 3 days. Shown are the mRNA amounts relative to the control animals as the means of 4-5 animals in each group ± SEM. B, Primary hepatocytes were isolated from wild-type and RIP140 null mice. Cells were treated with T0901317 or vehicle and glucose production was measured. C, PEPCK and G6Pase gene expression in primary hepatocytes form WT and RIP KO mice was determined by real-time PCR. D, Fao cells were depleted of RIP140 by siRNA (siRIP140). After 48 h control and siRIP140 cells were treated with T0901317 or vehicle alone for 12-16 h. PEPCK and G6Pase mRNA relative to control samples was measured by real-time PCR and represents the mean of 3 experiments ± SEM. *p<0.05, **p<0.01 versus animals or samples treated with vehicle alone.
Fig. 6. RIP140 binding to the PEPCK gene promoter
A, Coimmunopreciptiation assays were performed with nuclear extracts from Fao cells treated with 1μM T0901317 or vehicle alone for 2 h. B, Fao cells were treated with T0901317 or vehicle alone. Chromatin fragments were precipitated with the indicated antibodies and purified DNA was amplified by PCR using primers encompassing the LXREs in the promoter region of the PEPCK and G6Pase genes or a distal region of the PEPCK gene. The distal region of the PEPCK gene does not harbour a LXRE and was used as a control.
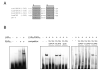
Fig. 7. Identification of a LXRE in the PEPCK gene promoter
A, Sequence alignment of putative LXREs in the rat, human and mouse PEPCK promoter and human FAS LXRE. B, Biotin-labeled PEPCK probe was incubated with LXRα and RXRα protein in the presence or absence of 10-50-fold molar excess of competitor oligonucleotides, as indicated. The resulting DNA-protein complexes were separated from the free probe in a polyacrylamide gel and detected using strepavidin conjugated to HRP.
Similar articles
- Molecular basis for repression of liver X receptor-mediated gene transcription by receptor-interacting protein 140.
Jakobsson T, Osman W, Gustafsson JA, Zilliacus J, Wärnmark A. Jakobsson T, et al. Biochem J. 2007 Jul 1;405(1):31-9. doi: 10.1042/BJ20070004. Biochem J. 2007. PMID: 17391100 Free PMC article. - Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue.
Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, Castrillo A, Wilpitz DC, Mangelsdorf DJ, Collins JL, Saez E, Tontonoz P. Laffitte BA, et al. Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5419-24. doi: 10.1073/pnas.0830671100. Epub 2003 Apr 15. Proc Natl Acad Sci U S A. 2003. PMID: 12697904 Free PMC article. - The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR.
Cha JY, Repa JJ. Cha JY, et al. J Biol Chem. 2007 Jan 5;282(1):743-51. doi: 10.1074/jbc.M605023200. Epub 2006 Nov 14. J Biol Chem. 2007. PMID: 17107947 - The metabolic coregulator RIP140: an update.
Fritah A, Christian M, Parker MG. Fritah A, et al. Am J Physiol Endocrinol Metab. 2010 Sep;299(3):E335-40. doi: 10.1152/ajpendo.00243.2010. Epub 2010 Jun 8. Am J Physiol Endocrinol Metab. 2010. PMID: 20530738 Review. - Functional crosstalk of CAR-LXR and ROR-LXR in drug metabolism and lipid metabolism.
Xiao L, Xie X, Zhai Y. Xiao L, et al. Adv Drug Deliv Rev. 2010 Oct 30;62(13):1316-21. doi: 10.1016/j.addr.2010.07.006. Epub 2010 Jul 24. Adv Drug Deliv Rev. 2010. PMID: 20659512 Review.
Cited by
- Behavioral stress reduces RIP140 expression in astrocyte and increases brain lipid accumulation.
Feng X, Lin YL, Wei LN. Feng X, et al. Brain Behav Immun. 2015 May;46:270-9. doi: 10.1016/j.bbi.2015.02.008. Epub 2015 Feb 16. Brain Behav Immun. 2015. PMID: 25697398 Free PMC article. - A liver-specific gene expression panel predicts the differentiation status of in vitro hepatocyte models.
Kim DS, Ryu JW, Son MY, Oh JH, Chung KS, Lee S, Lee JJ, Ahn JH, Min JS, Ahn J, Kang HM, Kim J, Jung CR, Kim NS, Cho HS. Kim DS, et al. Hepatology. 2017 Nov;66(5):1662-1674. doi: 10.1002/hep.29324. Epub 2017 Oct 3. Hepatology. 2017. PMID: 28640507 Free PMC article. - The nuclear receptor cofactor receptor-interacting protein 140 is a positive regulator of amphiregulin expression and cumulus cell-oocyte complex expansion in the mouse ovary.
Nautiyal J, Steel JH, Rosell MM, Nikolopoulou E, Lee K, Demayo FJ, White R, Richards JS, Parker MG. Nautiyal J, et al. Endocrinology. 2010 Jun;151(6):2923-32. doi: 10.1210/en.2010-0081. Epub 2010 Mar 22. Endocrinology. 2010. PMID: 20308529 Free PMC article. - Emerging role of liver X receptors in cardiac pathophysiology and heart failure.
Cannon MV, van Gilst WH, de Boer RA. Cannon MV, et al. Basic Res Cardiol. 2016 Jan;111(1):3. doi: 10.1007/s00395-015-0520-7. Epub 2015 Nov 26. Basic Res Cardiol. 2016. PMID: 26611207 Free PMC article. Review. - Nuclear receptors reverse McGarry's vicious cycle to insulin resistance.
Moore DD. Moore DD. Cell Metab. 2012 May 2;15(5):615-22. doi: 10.1016/j.cmet.2012.03.016. Cell Metab. 2012. PMID: 22560214 Free PMC article.
References
- Steffensen KR, Gustafsson JA. Putative metabolic effects of the liver X receptor (LXR) Diabetes. 2004;53(Suppl 1):S36–42. - PubMed
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. - PubMed
- Lazar MA, Willson TM. Sweet Dreams for LXR. Cell Metab. 2007;5:159–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 061930/WT_/Wellcome Trust/United Kingdom
- BB/C504327/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U120027537/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases