Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI - PubMed (original) (raw)
Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI
Miriam E Bucheli et al. RNA. 2007 Oct.
Abstract
Multiple steps in mRNA processing and transcription are coupled. Notably, the processing of mRNA 3' ends is linked to transcription termination by RNA polymerase II. Previously, we found that the yeast hnRNP protein Npl3 can negatively regulate 3' end mRNA formation and termination at the GAL1 gene. Here we show that overexpression of the Hrp1 or Rna14 subunits of the CF IA polyadenylation factor increases recognition of a weakened polyadenylation site. Genetic interactions of mutant alleles of NPL3 or HRP1 with RNA15 also indicate antagonism between these factors. Npl3 competes with Rna15 for binding to a polyadenylation precursor and inhibits cleavage and polyadenylation in vitro. These results suggest that an important function of hnRNP proteins is to ensure the fidelity of mRNA processing. Our results support a model in which balanced competition of Npl3 with mRNA processing factors may promote recognition of proper polyadenylation sites while suppressing cryptic sites.
Figures
FIGURE 1.
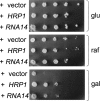
Overexpression of Hrp1 or Rna14 suppresses the sensitivity of gal10Δ56 to galactose. Serial 10-fold dilutions of a gal10Δ56 strain (CKY265), previously transformed with 2-μm plasmids containing either HRP1 or RNA14 as indicated, were spotted on SC-leu media supplemented with 2% glucose (glu), 2% raffinose (raf), or 2% raffinose/1% galactose (gal). Plates were incubated for 3 d (glu), 5 d (raf), or 7 d (gal) at 30°C. The gal10Δ56 strain transformed with a 2-μm vector is shown as control.
FIGURE 2.
HRP1 and RNA15 functions are essential for the _npl3_-mediated suppression of gal10Δ56. Serial 10-fold dilutions of strains with single or double mutations (hrp1-5, rna15-58, and npl3-120) in the gal10Δ56 background were spotted onto YPD or YPGal plates and grown for 2 d at 30°C. Alleles of hrp1-5 or rna15-58 with wild-type GAL10 are shown as controls for growth on YPGal.
FIGURE 3.
Npl3 competes with Rna15 for binding to the GAL7 3′UTR transcript. Recombinant Npl3, Hrp1, or Rna15/Rna14 complex were incubated with radioactively labeled RNA, UV cross-linked, and resolved in denaturing 10% SDS-PAGE gels. (A) Schematic diagram of the long transcript GAL7-1, including a U-rich, UA-repeat, and A-rich sequences upstream of the poly(A) site is at top. Representative UV-cross-link experiments are shown where increasing Npl3 is added to reactions containing Hrp1 or Rna15/Rna14. The graph below each gel shows quantitation for the average of three experiments, with error bars showing the standard deviation. Binding levels were calculated as a fraction relative to a reaction containing the highest concentration of the individual RNA-bound protein. (B) A UV cross-linking experiment similar to A was done where increasing levels of Npl3 were added to a reaction containing both Hrp1 and Rna15/Rna14 bound to the GAL7-1 RNA. (C) The same combination of proteins as that shown in B is shown with the GAL7-3 RNA. This template contains a deletion of the UA-repeat. (D) Same as B except for a truncated template (GAL7-4) where sequence upstream of the UA-repeat is deleted. (E) Same as B but with a minimal GAL7 (GAL7-1 min) sequence, which has an additional deletion that spans the region downstream of the poly(A) site.
FIGURE 4.
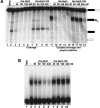
Wild-type and mutant Npl3 have distinct RNA cross-linking patterns. Npl3 and Npl3-120 were UV cross-linked to GAL7-1 RNA. Primer extension was then performed using a radiolabeled primer downstream of the poly(A) site; cross-links result in a block for reverse transcriptase and therefore give a band near the site of protein interaction. The RNA sequence, extrapolated from size markers, is shown next to the gels. (B) Same as A except a region (marked with an asterisk) further upstream of the poly(A) signal was expanded for analysis. Solid and dotted lines indicate PE stops for Npl3 and Npl3-120, respectively. (C) Coomassie staining showing the amount of each protein used in the cross-linking/primer extension analysis.
FIGURE 5.
Npl3 inhibits in vitro mRNA 3′ end processing. (A) In vitro mRNA 3′ end cleavage and polyadenylation assays. Yeast wild-type whole-cell extracts (30 μg; lanes 2_–_19) were incubated with radioactively labeled GAL7-1 RNA. Reactions were initiated with either dATP (for activation of cleavage but not polyadenylation, lanes 1_–_10) or ATP (allowing both cleavage and polyadenylation, lanes 11_–_19) for 20 min at 30°C. Whole-cell extract was supplemented with increasing concentrations of recombinant His6-Npl3 (lanes 3_–_6,7_–_10) or its mutant His6-Npl3–120 (lanes 7_–_10,16_–_19) as indicated before initiating the reaction. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager. The position of the precursor and product RNAs are marked schematically on the right. Unreacted precursor RNA is shown in lane 1, and lanes 2 and 11 show RNA incubated with whole-cell extract but no additional Npl3. (B) Poly(A) addition assays. The assays were performed as in A except that precleaved GAL7-9 RNA (lane 1) was used instead as the RNA precursor.
Similar articles
- Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation.
Dermody JL, Dreyfuss JM, Villén J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Dermody JL, et al. PLoS One. 2008 Sep 26;3(9):e3273. doi: 10.1371/journal.pone.0003273. PLoS One. 2008. PMID: 18818768 Free PMC article. - Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I.
Gross S, Moore C. Gross S, et al. Proc Natl Acad Sci U S A. 2001 May 22;98(11):6080-5. doi: 10.1073/pnas.101046598. Epub 2001 May 8. Proc Natl Acad Sci U S A. 2001. PMID: 11344258 Free PMC article. - Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3'-end formation.
Gross S, Moore CL. Gross S, et al. Mol Cell Biol. 2001 Dec;21(23):8045-55. doi: 10.1128/MCB.21.23.8045-8055.2001. Mol Cell Biol. 2001. PMID: 11689695 Free PMC article. - Architectural and functional details of CF IA proteins involved in yeast 3'-end pre-mRNA processing and its significance for eukaryotes: A concise review.
Kaur M, Sharma A, Singh G, Kumar S, Barnwal RP. Kaur M, et al. Int J Biol Macromol. 2021 Dec 15;193(Pt A):387-400. doi: 10.1016/j.ijbiomac.2021.10.129. Epub 2021 Oct 23. Int J Biol Macromol. 2021. PMID: 34699898 Review. - The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5' capping and splicing.
Yang Q, Gilmartin GM, Doublié S. Yang Q, et al. RNA Biol. 2011 Sep-Oct;8(5):748-53. doi: 10.4161/rna.8.5.16040. Epub 2011 Sep 1. RNA Biol. 2011. PMID: 21881408 Free PMC article. Review.
Cited by
- Transcription termination by nuclear RNA polymerases.
Richard P, Manley JL. Richard P, et al. Genes Dev. 2009 Jun 1;23(11):1247-69. doi: 10.1101/gad.1792809. Genes Dev. 2009. PMID: 19487567 Free PMC article. - Interactions affected by arginine methylation in the yeast protein-protein interaction network.
Erce MA, Abeygunawardena D, Low JK, Hart-Smith G, Wilkins MR. Erce MA, et al. Mol Cell Proteomics. 2013 Nov;12(11):3184-98. doi: 10.1074/mcp.M113.031500. Epub 2013 Aug 5. Mol Cell Proteomics. 2013. PMID: 23918811 Free PMC article. - Novel protein-protein contacts facilitate mRNA 3'-processing signal recognition by Rna15 and Hrp1.
Leeper TC, Qu X, Lu C, Moore C, Varani G. Leeper TC, et al. J Mol Biol. 2010 Aug 20;401(3):334-49. doi: 10.1016/j.jmb.2010.06.032. Epub 2010 Jun 19. J Mol Biol. 2010. PMID: 20600122 Free PMC article. - To the pore and through the pore: a story of mRNA export kinetics.
Oeffinger M, Zenklusen D. Oeffinger M, et al. Biochim Biophys Acta. 2012 Jun;1819(6):494-506. doi: 10.1016/j.bbagrm.2012.02.011. Epub 2012 Feb 22. Biochim Biophys Acta. 2012. PMID: 22387213 Free PMC article. Review. - T helper cells exhibit a dynamic and reversible 3'-UTR landscape.
Seyres D, Gorka O, Schmidt R, Marone R, Zavolan M, Jeker LT. Seyres D, et al. RNA. 2024 Mar 18;30(4):418-434. doi: 10.1261/rna.079897.123. RNA. 2024. PMID: 38302256 Free PMC article.
References
- Buratowski, S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM056663/GM/NIGMS NIH HHS/United States
- K01 CA115515/CA/NCI NIH HHS/United States
- R01 GM056663-09/GM/NIGMS NIH HHS/United States
- 1K01CA115515-01A1/CA/NCI NIH HHS/United States
- R01 GM068887/GM/NIGMS NIH HHS/United States
- GM56663/GM/NIGMS NIH HHS/United States
- GM68887/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous