Drosophila overexpressing parkin R275W mutant exhibits dopaminergic neuron degeneration and mitochondrial abnormalities - PubMed (original) (raw)
Comparative Study
Drosophila overexpressing parkin R275W mutant exhibits dopaminergic neuron degeneration and mitochondrial abnormalities
Cheng Wang et al. J Neurosci. 2007.
Abstract
Mutations in the parkin gene are a predominant cause of familial parkinsonism. Although initially described as a recessive disorder, emerging evidence suggest that single parkin mutations alone may confer increased susceptibility to Parkinson's disease. To better understand the effects of parkin mutations in vivo, we generated transgenic Drosophila overexpressing two human parkin missense mutants, R275W and G328E. Transgenic flies that overexpress R275W, but not wild-type or G328E, human parkin display an age-dependent degeneration of specific dopaminergic neuronal clusters and concomitant locomotor deficits that accelerate with age or in response to rotenone treatment. Furthermore, R275W mutant flies also exhibit prominent mitochondrial abnormalities in their flight muscles. Interestingly, these defects caused by the expression of human R275W parkin are highly similar to those triggered by the loss of endogenous parkin in parkin null flies. Together, our results provide the first in vivo evidence demonstrating that parkin R275W mutant expression mediates pathogenic outcomes and suggest the interesting possibility that select parkin mutations may directly exert neurotoxicity in vivo.
Figures
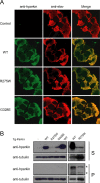
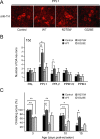
Figure 1.
Pan-neuronal expression of parkin mutants in transgenic Drosophila. A, Anti-human parkin (hparkin) (green) and anti-elav (red) immunostaining of whole-mount adult brains derived from 20-d-old transgenic flies expressing normal or mutant parkin species, as indicated. B, Left, Anti-parkin immunoblot of detergent-soluble (S) and detergent-insoluble (P) fractionated brain lysates derived from 20-d-old wild-type (WT) or transgenic (Tg) adult fly brains. Right, As above except that three times more brain lysates from R275W flies were loaded relative to those from wild-type parkin-expressing flies. Membranes were stripped and reprobed with anti-tubulin to reflect loading variations. Asterisks denotes nonspecific bands (genotype: elav-GAL4/+; UAS-hParkin/+, control elav-GAL4/+)
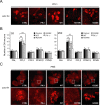
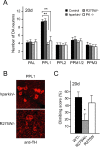
Figure 2.
Expression of parkin R275W mutant in flies promotes dopaminergic neuronal degeneration in select clusters. A, Representative confocal microscopy images showing TH-positive (red) dopaminergic neurons in the PPL1 cluster of 20-d-old control (CTRL) and parkin null flies (PK−/−) as well as in transgenic flies expressing wild-type (WT) or mutant parkin species, as indicated. B, Bar graph showing the number of dopaminergic (DA) neurons in different clusters of the various fly species, as depicted by different shades shown in the figure (*p < 0.05, **p < 0.01, Student's t test; n = 10). C, Representative confocal microscopy images showing TH-positive (red) dopaminergic neurons in the PAM cluster of 20-d-old control and parkin null flies (PK−/−) as well as in transgenic flies expressing normal or mutant parkin species, as indicated. PAM cluster in top row (boxed) are shown at higher magnification in corresponding bottom row (genotype: Ddc-Gal4/+; UAS-hParkin/+, control Ddc-Gal4/+)
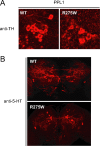
Figure 3.
Selective dopaminergic neuronal loss in R275W mutant flies. A, B, Representative confocal microscopy images showing TH-positive (red) dopaminergic neurons in the PPL1 cluster (A) and 5-HT-positive neurons (red) of 20-d-old transgenic flies expressing wild-type (WT) or R275W mutant parkin under the _elav_-GAL4 driver (B), as indicated (genotype: elav-GAL4/+; UAS-hParkin/+).
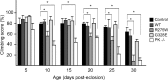
Figure 4.
Parkin null and transgenic parkin R275W flies exhibit impaired climbing ability. Bar graph showing the percentage of flies at different days after eclosion that reached the top of assay column after 1 min. The various fly strains examined are depicted by different shades shown in the figure (*p < 0.05, **p < 0.01, Student's t test; n = 20) (genotype: Ddc-Gal4/+; UAS-hParkin/+, control Ddc-Gal4/+). WT, Wild type.
Figure 5.
Exposure to rotenone accelerates PPL1 dopaminergic neurodegeneration and locomotor deficits in transgenic parkin R275W mutant flies. A, Representative confocal microscopy images showing TH-positive (red) dopaminergic neurons in the PPL1 cluster of 15-d-old, rotenone-treated, transgenic flies expressing wild-type (WT) or mutant parkin species, as indicated. B, Bar graph showing the number of dopaminergic (DA) neurons in different clusters of the various fly species treated with rotenone, as depicted by different shades shown in the figure (*p < 0.05, **_p_ < 0.01, Student's _t_ test; _n_ = 10). **_C_**, Bar graph showing the climbing scores of the various rotenone-treated flies at different days after eclosion, as depicted by different shades shown in the figure (*_p_ < 0.05, **_p_ < 0.01 Student's _t_ test; _n_ > 20) (genotype: Ddc-Gal4/+; UAS-hParkin/+, control Ddc-Gal4/+).
Figure 6.
Overexpression of wild-type and R275W parkin in parkin null flies exert different effects on dopaminergic neuronal survivability. A, Bar graph showing the number of dopaminergic (DA) neurons in different clusters of 20-d-old hparkin/− and R275W/− flies, as depicted by different shades shown in the figure. Data for control and parkin null flies were derived from Figure 2 (*p < 0.05, **p < 0.01, Student's t test; n = 10). B, Representative confocal microscopy images showing TH-positive (red) dopaminergic neurons in the PPL1 cluster of 20-d-old old hparkin/− and R275W/− flies, as indicated. C, Bar graph showing the climbing scores of the various mutant flies, as indicated, at 20 d after eclosion (*p < 0.05, **p < 0.01, Student's t test; n = 20) (genotype: UAS-hParkin, park1/park1, Ddc-Gal4, control Ddc-Gal4/+).
Figure 7.
Mitochondrial defects in parkin R275W mutant flies. TEM analysis of indirect flight muscles of 20-d-old flies expressing wild-type human parkin (WT) (A, B), R275W (C, D), G328E (E, F), or lacking endogenous parkin (PK−/−) (G, H) (genotype: 24B-Gal4/+; UAS-hParkin/+). The same analysis is performed in 2-d-old parkin null flies (I, J) or those expressing wild-type human parkin (K, L) or R275W (M, N) against parkin null background (genotype: UAS-hParkin, park1/park1, 24B-Gal4). Scale bars: A, C, E, G, I, K, M, 1 μ
m
; B, D, F, H, J, L, N, 0.5 μ
m
.
Similar articles
- A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine.
Sang TK, Chang HY, Lawless GM, Ratnaparkhi A, Mee L, Ackerson LC, Maidment NT, Krantz DE, Jackson GR. Sang TK, et al. J Neurosci. 2007 Jan 31;27(5):981-92. doi: 10.1523/JNEUROSCI.4810-06.2007. J Neurosci. 2007. PMID: 17267552 Free PMC article. - PARIS induced defects in mitochondrial biogenesis drive dopamine neuron loss under conditions of parkin or PINK1 deficiency.
Pirooznia SK, Yuan C, Khan MR, Karuppagounder SS, Wang L, Xiong Y, Kang SU, Lee Y, Dawson VL, Dawson TM. Pirooznia SK, et al. Mol Neurodegener. 2020 Mar 5;15(1):17. doi: 10.1186/s13024-020-00363-x. Mol Neurodegener. 2020. PMID: 32138754 Free PMC article. - Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila.
Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Ng CH, et al. J Neurosci. 2009 Sep 9;29(36):11257-62. doi: 10.1523/JNEUROSCI.2375-09.2009. J Neurosci. 2009. PMID: 19741132 Free PMC article. - PINK1-Parkin signaling in Parkinson's disease: Lessons from Drosophila.
Imai Y. Imai Y. Neurosci Res. 2020 Oct;159:40-46. doi: 10.1016/j.neures.2020.01.016. Epub 2020 Feb 6. Neurosci Res. 2020. PMID: 32035987 Review. - The synaptic function of parkin.
Sassone J, Serratto G, Valtorta F, Silani V, Passafaro M, Ciammola A. Sassone J, et al. Brain. 2017 Sep 1;140(9):2265-2272. doi: 10.1093/brain/awx006. Brain. 2017. PMID: 28335015 Review.
Cited by
- Optic atrophy 1 mediates mitochondria remodeling and dopaminergic neurodegeneration linked to complex I deficiency.
Ramonet D, Perier C, Recasens A, Dehay B, Bové J, Costa V, Scorrano L, Vila M. Ramonet D, et al. Cell Death Differ. 2013 Jan;20(1):77-85. doi: 10.1038/cdd.2012.95. Epub 2012 Aug 3. Cell Death Differ. 2013. PMID: 22858546 Free PMC article. - Drosophila models of Parkinson's disease: discovering relevant pathways and novel therapeutic strategies.
Muñoz-Soriano V, Paricio N. Muñoz-Soriano V, et al. Parkinsons Dis. 2011 Mar 3;2011:520640. doi: 10.4061/2011/520640. Parkinsons Dis. 2011. PMID: 21512585 Free PMC article. - Analysis of α-syn and parkin interaction in mediating neuronal death in Drosophila model of Parkinson's disease.
Narwal S, Singh A, Tare M. Narwal S, et al. Front Cell Neurosci. 2024 Jan 4;17:1295805. doi: 10.3389/fncel.2023.1295805. eCollection 2023. Front Cell Neurosci. 2024. PMID: 38239290 Free PMC article. - LRRK2 mutations and neurotoxicant susceptibility.
Lee JW, Cannon JR. Lee JW, et al. Exp Biol Med (Maywood). 2015 Jun;240(6):752-9. doi: 10.1177/1535370215579162. Epub 2015 Apr 16. Exp Biol Med (Maywood). 2015. PMID: 25888648 Free PMC article. Review. - α-Arbutin Protects Against Parkinson's Disease-Associated Mitochondrial Dysfunction In Vitro and In Vivo.
Ding Y, Kong D, Zhou T, Yang ND, Xin C, Xu J, Wang Q, Zhang H, Wu Q, Lu X, Lim K, Ma B, Zhang C, Li L, Huang W. Ding Y, et al. Neuromolecular Med. 2020 Mar;22(1):56-67. doi: 10.1007/s12017-019-08562-6. Epub 2019 Aug 10. Neuromolecular Med. 2020. PMID: 31401719
References
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. - PubMed
- Clark LN, Afridi S, Karlins E, Wang Y, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, Waters C, Ford B, Frucht S, Ottman R, Marder K. Case-control study of the parkin gene in early-onset Parkinson disease. Arch Neurol. 2006;63:548–552. - PubMed
- Cookson MR. Neurodegeneration: how does parkin prevent Parkinson's disease? Curr Biol. 2003;13:R522–R524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases