DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA - PubMed (original) (raw)
. 2007 Aug 9;448(7154):714-7.
doi: 10.1038/nature05987.
Chen Qiu, Emily Bernstein, Keqin Li, Da Jia, Zhe Yang, Hediye Erdjument-Bromage, Paul Tempst, Shau-Ping Lin, C David Allis, Xiaodong Cheng, Timothy H Bestor
Affiliations
- PMID: 17687327
- PMCID: PMC2650820
- DOI: 10.1038/nature05987
DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA
Steen K T Ooi et al. Nature. 2007.
Abstract
Mammals use DNA methylation for the heritable silencing of retrotransposons and imprinted genes and for the inactivation of the X chromosome in females. The establishment of patterns of DNA methylation during gametogenesis depends in part on DNMT3L, an enzymatically inactive regulatory factor that is related in sequence to the DNA methyltransferases DNMT3A and DNMT3B. The main proteins that interact in vivo with the product of an epitope-tagged allele of the endogenous Dnmt3L gene were identified by mass spectrometry as DNMT3A2, DNMT3B and the four core histones. Peptide interaction assays showed that DNMT3L specifically interacts with the extreme amino terminus of histone H3; this interaction was strongly inhibited by methylation at lysine 4 of histone H3 but was insensitive to modifications at other positions. Crystallographic studies of human DNMT3L showed that the protein has a carboxy-terminal methyltransferase-like domain and an N-terminal cysteine-rich domain. Cocrystallization of DNMT3L with the tail of histone H3 revealed that the tail bound to the cysteine-rich domain of DNMT3L, and substitution of key residues in the binding site eliminated the H3 tail-DNMT3L interaction. These data indicate that DNMT3L recognizes histone H3 tails that are unmethylated at lysine 4 and induces de novo DNA methylation by recruitment or activation of DNMT3A2.
Figures
Figure 1. Generation of epitope-tagged Dnmt3L locus and DNMT3L protein interaction screen in ES cells
a, Targeting of the endogenous mouse Dnmt3L locus with a replacement cassette that introduces an N-terminal His6-FLAG epitope tag. The tag was introduced immediately after the first four amino acids of DNMT3L (Supplementary Fig. S1). The neo resistance cassette was deleted by transient exposure to Cre recombinase. Mice homozygous for the Dnmt3LTag allele are fertile and were used to generate homozygous Dnmt3LTag/Tag ES cells. The southern blot on the right shows that homologous recombination and Cre-mediated excision of the marker occurred as predicted. b, Protein interaction screen in wild-type and Dnmt3LTag/Tag ES cells by FLAG immunoprecipitation. Coomassie-stained bands were subjected to MALDI-reTOF and mass spectra were screened against a non-redundant protein database to identify interacting proteins. The protein band marked by the asterisk is actin, a common contaminant in anti-FLAG immunoprecipitation.
Figure 2. Interaction of DNMT3L with the N terminus of histone H3 is abolished by methylation of H3 lysine 4
a, Peptide interaction assays with unmodified histone tails as indicated, incubated with full-length human DNMT3L. DNMT3L interacts only with the N-terminal tail of H3 (left), and this interaction requires only the first seven amino acids of H3 (right). b, Peptide interaction assays with DNMT3L and modified histone tails as indicated. DNMT3L bound only to the H3 tail that is unmodified at lysine 4. c, WDR5 (WD repeat domain 5) and the chromodomain of HP-1α (heterochromatin protein 1α) were used as controls for peptide binding specificity. d, Dissociation constants as determined by fluorescence polarization with C-terminal fluoresceinated peptides. _K_d values are shown on the right. The 18-fold increase in _K_d caused by monomethylation of H3 lysine 4 prevented detection of DNMT3L binding in b. e, Histones associated with DNMT3L in vivo are depleted in H3 that is trimethylated at lysine 4. Nuclear extract was treated with micrococcal nuclease before immunoprecipitation followed by separation of proteins by SDS-polyacrylamide gel electrophoresis. Immunoblot was performed with the antibodies indicated. A specific depletion in H3 trimethylated at lysine 4 can be seen in the fourth immunoblot from the top.
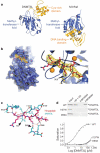
Figure 3. Structure of DNMT3L and mode of recognition of histone H3 unmethylated at lysine 4
a, DNMT3L contains a classical methyltransferase fold as well as a cysteine-rich region organized around three zinc ions (indicated in yellow; zinc ions in orange). The structure of DNMT3L alone was determined to a resolution of 3.29 Å. M._Hha_I is a bacterial DNA cytosine-5 methyltransferase that contains a classical methyltransferase fold. b, Structure of DNMT3L after incorporation of the H3 N-terminal tail into the DNMT3L crystal by soaking with peptide. The omit electron density (orange) is contoured at 4.0σ. At the resolution of 3.69 Å, the electron density map alone does not provide an unambiguous position of the bound peptide. The model shown here is based on stereochemical principles and on the strong structural similarity with BHC80, which also binds specifically to histone H3 unmethylated at lysine 4 (Supplementary Fig. S2 and ref. 14). Only the N-terminal seven amino acids of the 24-amino-acid peptide were structured. The side chain of R2 was also disordered in the crystal. c, Detailed interaction between the H3 N terminus (amino acids numbered in red) and DNMT3L (numbered in black). The dashed lines indicate potential interactions between amino-acid side chains. H3 lysine 4 makes contacts with the carboxylates of DNMT3L D90 and D88, and methylation of lysine 4 will occlude these interactions. d, Mutagenesis of residues in the peptide binding site of DNMT3L abolished binding. Arrowheads in c indicate positions of I107W and D90A mutations. Each mutation abolished the interaction of DNMT3L with the unmethylated N-terminal peptide as shown by binding assay (top) and fluorescence polarization (bottom). _K_d for binding to the wild type was 2.1 μM and for each of the two mutated proteins was >500 μM.
Similar articles
- Structural insight into autoinhibition and histone H3-induced activation of DNMT3A.
Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Guo X, et al. Nature. 2015 Jan 29;517(7536):640-4. doi: 10.1038/nature13899. Epub 2014 Nov 10. Nature. 2015. PMID: 25383530 - Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Jia D, et al. Nature. 2007 Sep 13;449(7159):248-51. doi: 10.1038/nature06146. Epub 2007 Aug 22. Nature. 2007. PMID: 17713477 Free PMC article. - Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail.
Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Zhang Y, et al. Nucleic Acids Res. 2010 Jul;38(13):4246-53. doi: 10.1093/nar/gkq147. Epub 2010 Mar 11. Nucleic Acids Res. 2010. PMID: 20223770 Free PMC article. - Functions of DNA methyltransferase 3-like in germ cells and beyond.
Liao HF, Tai KY, Chen WS, Cheng LC, Ho HN, Lin SP. Liao HF, et al. Biol Cell. 2012 Oct;104(10):571-87. doi: 10.1111/boc.201100109. Epub 2012 Jul 10. Biol Cell. 2012. PMID: 22671959 Review. - Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases.
Tajima S, Suetake I, Takeshita K, Nakagawa A, Kimura H. Tajima S, et al. Adv Exp Med Biol. 2016;945:63-86. doi: 10.1007/978-3-319-43624-1_4. Adv Exp Med Biol. 2016. PMID: 27826835 Review.
Cited by
- Gene regulation by the act of long non-coding RNA transcription.
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Kornienko AE, et al. BMC Biol. 2013 May 30;11:59. doi: 10.1186/1741-7007-11-59. BMC Biol. 2013. PMID: 23721193 Free PMC article. Review. - The promise and failures of epigenetic therapies for cancer treatment.
Bojang P Jr, Ramos KS. Bojang P Jr, et al. Cancer Treat Rev. 2014 Feb;40(1):153-69. doi: 10.1016/j.ctrv.2013.05.009. Epub 2013 Jul 5. Cancer Treat Rev. 2014. PMID: 23831234 Free PMC article. Review. - H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation.
Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. Yang SM, et al. Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1708-13. doi: 10.1073/pnas.1213266110. Epub 2013 Jan 9. Proc Natl Acad Sci U S A. 2013. PMID: 23302691 Free PMC article. - Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions.
Chen J, Horton J, Sagum C, Zhou J, Cheng X, Bedford MT. Chen J, et al. Biochem J. 2021 May 28;478(10):1943-1958. doi: 10.1042/BCJ20210203. Biochem J. 2021. PMID: 33969871 Free PMC article. - Therapeutic modulation of gene expression in the disease state: Treatment strategies and approaches for the development of next-generation of the epigenetic drugs.
Rittiner J, Cumaran M, Malhotra S, Kantor B. Rittiner J, et al. Front Bioeng Biotechnol. 2022 Oct 17;10:1035543. doi: 10.3389/fbioe.2022.1035543. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36324900 Free PMC article. Review.
References
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. - PubMed
- Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J.Biol. Chem. 2002;277:38746–38754. - PubMed
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials