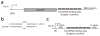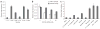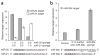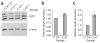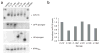MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells - PubMed (original) (raw)
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
Margaret S Ebert et al. Nat Methods. 2007 Sep.
Abstract
MicroRNAs are predicted to regulate thousands of mammalian genes, but relatively few targets have been experimentally validated and few microRNA loss-of-function phenotypes have been assigned. As an alternative to chemically modified antisense oligonucleotides, we developed microRNA inhibitors that can be expressed in cells, as RNAs produced from transgenes. Termed 'microRNA sponges', these competitive inhibitors are transcripts expressed from strong promoters, containing multiple, tandem binding sites to a microRNA of interest. When vectors encoding these sponges are transiently transfected into cultured cells, sponges derepress microRNA targets at least as strongly as chemically modified antisense oligonucleotides. They specifically inhibit microRNAs with a complementary heptameric seed, such that a single sponge can be used to block an entire microRNA seed family. RNA polymerase II promoter (Pol II)-driven sponges contain a fluorescence reporter gene for identification and sorting of sponge-treated cells. We envision the use of stably expressed sponges in animal models of disease and development.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Figures
Figure 1
Design of microRNA sponges. (a) We constructed GFP sponges by inserting multiple microRNA binding sites into the 3′ UTR of a 2-h destabilized GFP reporter gene driven by the CMV promoter. (b) The imperfect pairing between a microRNA and a sponge with bulged binding sites is diagrammed for miR-21. We designed sponges with a bulge to protect against endonucleolytic cleavage by Argonaute 2. (c) We constructed U6 sponges by subcloning the microRNA binding site region into a vector containing a U6 snRNA promoter with 5′ and 3′ stem-loop elements.
Figure 2
Efficacy of microRNA sponges. (a–c) RLuc activity relative to firefly luciferase activity was assayed in 293T cells 24 h after transfection with RLuc microRNA target reporters, firefly luciferase transfection control and microRNA sponge plasmids. An RLuc target regulated by 7 miR-20 sites was derepressed by GFP sponges and U6 sponges with bulged or perfect binding sites for miR-20 (a). C, CMV sponge; U, U6 sponge. CX, CXCR4 control; 20b, 7 bulged miR-20 sites; 20pf, two perfect miR-20 sites. Bars represent the expression of the miR-20 target relative to an untargeted control reporter. We measured an artificial CXCR4 target reporter with a single bulged binding site in the presence of a control GFP sponge against miR-21 (miR-21 sponge) or a GFP sponge containing seven CXCR4 binding sites (CXCR4 sponge; b). We transfected cells with 20 nM antisense oligonucleotide (2′ _O_-methyl 20 or LNA 20) or with the CMV bulged sponge against miR-20 (sponge 20; c). Negative controls; mock (no oligonucleotides or sponges), 2′ _O_-methyl against miR-30, LNA against miR-122, CXCR4 sponge. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
Figure 3
Specificity of microRNA sponges. (a) We assayed RLuc activity relative to firefly luciferase activity in HeLa cells 24 h after transfection with RLuc microRNA target reporters, firefly luciferase transfection control and microRNA sponge plasmids. Targets of miR-20 and miR-21 are specifically derepressed by the corresponding GFP sponge. Bars are normalized to the relative RLuc units of samples treated with the CXCR4 control sponge. (b) We assayed a perfect target reporter of miR-30c in HeLa cells transfected with oligonucleotide or sponge inhibitors of miR-30e. Controls: 2’ _O_-methyl anti–miR-181, CXCR4 sponge. MicroRNA sequences below show the heptameric seed sequence in bold, with nucleotide differences between the two family members underlined. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
Figure 4
Validation of microRNA targets. (a) We assayed 293T cells transfected with GFP sponges against miR-18, miR-20 or the CXCR4 control by western blot 48 h after transfection. The increase in endogenous E2F1 upon inhibition of miR-18 or miR-20 is shown relative to the control samples loaded at indicated amounts; E2F1 is the ~60 kDa band indicated; the other bands are nonspecific (top). β-actin loading control (bottom). (b) We assayed 293T cells transfected with an RLuc reporter fused to a fragment of the E2F1 UTR spanning two miR-20 sites, firefly luciferase and GFP sponges. Bars represent RLuc units relative to firefly luciferase units. (c) We assayed RLuc activity relative to firefly luciferase activity in 293T cells transfected with an RLuc reporter fused to a fragment of the CD69 UTR containing a predicted miR-20 binding site, firefly luciferase and GFP sponges. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
Figure 5
Effect of sponges on microRNA levels. (a) We transfected 293T cells with sponge plasmids and collected total RNA for northern blot analysis 48 h later. We probed the blot for miR-16 (top; 24-h exposure), then stripped the blot and reprobed it for GFP mRNA, U6 sponge RNA (both 24-h exposure) and a tRNA loading control (3-h exposure; bottom). (b) Quantitation of miR-16 relative to tRNA for each sponge-treated sample. We performed northern blots for miR-16 and miR-20 >10 times in 293T and HeLa cells, and show results from a representative blot.
Comment in
- Soaking up small RNAs.
Hammond SM. Hammond SM. Nat Methods. 2007 Sep;4(9):694-5. doi: 10.1038/nmeth0907-694. Nat Methods. 2007. PMID: 17762876 No abstract available.
Similar articles
- Optimization of a microRNA expression vector for function analysis of microRNA.
Furukawa N, Sakurai F, Katayama K, Seki N, Kawabata K, Mizuguchi H. Furukawa N, et al. J Control Release. 2011 Feb 28;150(1):94-101. doi: 10.1016/j.jconrel.2010.12.001. Epub 2010 Dec 10. J Control Release. 2011. PMID: 21146569 - MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development.
Boutz PL, Chawla G, Stoilov P, Black DL. Boutz PL, et al. Genes Dev. 2007 Jan 1;21(1):71-84. doi: 10.1101/gad.1500707. Genes Dev. 2007. PMID: 17210790 Free PMC article. - Gene Reporter Assays to Study miRNA Function.
Abel Y, Rederstorff M. Abel Y, et al. Methods Mol Biol. 2021;2300:119-131. doi: 10.1007/978-1-0716-1386-3_12. Methods Mol Biol. 2021. PMID: 33792877 - Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells.
Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Tay FC, et al. Adv Drug Deliv Rev. 2015 Jan;81:117-27. doi: 10.1016/j.addr.2014.05.010. Epub 2014 May 22. Adv Drug Deliv Rev. 2015. PMID: 24859534 Review. - Emerging roles for natural microRNA sponges.
Ebert MS, Sharp PA. Ebert MS, et al. Curr Biol. 2010 Oct 12;20(19):R858-61. doi: 10.1016/j.cub.2010.08.052. Curr Biol. 2010. PMID: 20937476 Free PMC article. Review.
Cited by
- LncRNA BRE-AS1 regulates the JAK2/STAT3-mediated inflammatory activation via the miR-30b-5p/SOC3 axis in THP-1 cells.
Shin JJ, Suk K, Lee WH. Shin JJ, et al. Sci Rep. 2024 Oct 28;14(1):25726. doi: 10.1038/s41598-024-77265-1. Sci Rep. 2024. PMID: 39468152 Free PMC article. - MicroRNA delivery for regenerative medicine.
Peng B, Chen Y, Leong KW. Peng B, et al. Adv Drug Deliv Rev. 2015 Jul 1;88:108-22. doi: 10.1016/j.addr.2015.05.014. Epub 2015 May 27. Adv Drug Deliv Rev. 2015. PMID: 26024978 Free PMC article. Review. - The code of non-coding RNAs in lung fibrosis.
Cui H, Xie N, Thannickal VJ, Liu G. Cui H, et al. Cell Mol Life Sci. 2015 Sep;72(18):3507-19. doi: 10.1007/s00018-015-1939-6. Epub 2015 May 31. Cell Mol Life Sci. 2015. PMID: 26026420 Free PMC article. Review. - Oligonucleotide-Based Therapeutics for STAT3 Targeting in Cancer-Drug Carriers Matter.
Molenda S, Sikorska A, Florczak A, Lorenc P, Dams-Kozlowska H. Molenda S, et al. Cancers (Basel). 2023 Nov 29;15(23):5647. doi: 10.3390/cancers15235647. Cancers (Basel). 2023. PMID: 38067351 Free PMC article. Review. - Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria.
Bossi L, Figueroa-Bossi N. Bossi L, et al. Nat Rev Microbiol. 2016 Dec;14(12):775-784. doi: 10.1038/nrmicro.2016.129. Epub 2016 Sep 19. Nat Rev Microbiol. 2016. PMID: 27640758 Review.
References
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
- Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA014051/CA/NCI NIH HHS/United States
- U19-AI056900/AI/NIAID NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- U54 CA112967/CA/NCI NIH HHS/United States
- U19 AI056900/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials