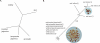Global dissemination of a single mutation conferring white pericarp in rice - PubMed (original) (raw)
Global dissemination of a single mutation conferring white pericarp in rice
Megan T Sweeney et al. PLoS Genet. 2007 Aug.
Abstract
Here we report that the change from the red seeds of wild rice to the white seeds of cultivated rice (Oryza sativa) resulted from the strong selective sweep of a single mutation, a frame-shift deletion within the Rc gene that is found in 97.9% of white rice varieties today. A second mutation, also within Rc, is present in less than 3% of white accessions surveyed. Haplotype analysis revealed that the predominant mutation originated in the japonica subspecies and crossed both geographic and sterility barriers to move into the indica subspecies. A little less than one Mb of japonica DNA hitchhiked with the rc allele into most indica varieties, suggesting that other linked domestication alleles may have been transferred from japonica to indica along with white pericarp color. Our finding provides evidence of active cultural exchange among ancient farmers over the course of rice domestication coupled with very strong, positive selection for a single white allele in both subspecies of O. sativa.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
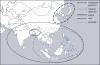
Figure 1. Current Distribution of the Five Major Subpopulations of Rice in Asia
For details about the geographical distribution of accessions used in this study, see Table S1.
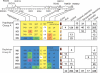
Figure 2. Haplotype Data across the Rc Gene
The gene model for Rc, containing seven exons and comprising a region of approximately 8 kb, is shown horizontally along the top; arrows on the gene model and promoter region of Rc indicate positions of markers: RID-number markers amplify rice insertion/deletion polymorhisms, RM-number markers amplify rice microsatellite polymorphisms, and the RS-number marker detects a rice SNP. Physical positions along the rice Chromosome 7 pseudomolecule are indicated by the ruler below the gene model. In the tables below, allele sizes for RID marker loci are given in bp and allele sizes for RM markers show the number of repeats at each microsatellite locus. The number of varieties in each subpopulation that carry each haplotype is provided in boxed squares in the table to the right. Haplotypes found in varieties with red pericarp are shaded red.
Figure 3. Comparison of Rc Gene and Rice Genome-Wide Phylogenetic Trees
Sequence alignment adjusted manually in MacClade 4.05. A heuristic parsimony search was conducted on the data matrix, excluding gapped characters, with TBR branch swapping. This identified a single tree at 53 steps, with a consistency index of 0.98 (0.97 without autapomorphies) and a retention index of 0.99. Bootstrap support was estimated using 100 replications of the same search strategy. Pericarp color and subpopulation of each variety sequenced is labeled.
Figure 4. Japonica Introgressions in White Indica
Arrows indicate the location of markers on the physical map of Chromosome 7, numbers are in Mb. F ST values for red indica versus japonica are plotted across the chromosome using diamonds, white indica versus japonica are plotted using squares. The expanded region shows a detailed view of the 1-Mb window around the Rc gene. All varieties genotyped are white indicas containing the 14-bp deletion. DNA segments containing alleles of japonica ancestry (in the indica background) are shown in white, segments containing alleles of indica ancestry are shown in black, and segments containing a breakpoint between alleles of japonica and indica ancestry are shown in grey. Locations of the Rc and Hsh genes indicated by brackets.
Similar articles
- Little White Lies: Pericarp Color Provides Insights into the Origins and Evolution of Southeast Asian Weedy Rice.
Cui Y, Song BK, Li LF, Li YL, Huang Z, Caicedo AL, Jia Y, Olsen KM. Cui Y, et al. G3 (Bethesda). 2016 Dec 7;6(12):4105-4114. doi: 10.1534/g3.116.035881. G3 (Bethesda). 2016. PMID: 27729434 Free PMC article. - Pericarp color and haplotype diversity in weedy rice (O. sativa f. spontanea) from Thailand.
Prathepha P. Prathepha P. Pak J Biol Sci. 2009 Aug 1;12(15):1075-9. doi: 10.3923/pjbs.2009.1075.1079. Pak J Biol Sci. 2009. PMID: 19943464 - LABA1, a Domestication Gene Associated with Long, Barbed Awns in Wild Rice.
Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P, Cai H, McCouch SR, Sun C. Hua L, et al. Plant Cell. 2015 Jul;27(7):1875-88. doi: 10.1105/tpc.15.00260. Epub 2015 Jun 16. Plant Cell. 2015. PMID: 26082172 Free PMC article. - The complex history of the domestication of rice.
Sweeney M, McCouch S. Sweeney M, et al. Ann Bot. 2007 Nov;100(5):951-7. doi: 10.1093/aob/mcm128. Epub 2007 Jul 6. Ann Bot. 2007. PMID: 17617555 Free PMC article. Review. - New insights into the history of rice domestication.
Kovach MJ, Sweeney MT, McCouch SR. Kovach MJ, et al. Trends Genet. 2007 Nov;23(11):578-87. doi: 10.1016/j.tig.2007.08.012. Epub 2007 Oct 25. Trends Genet. 2007. PMID: 17963977 Review.
Cited by
- Diversity in the complexity of phosphate starvation transcriptomes among rice cultivars based on RNA-Seq profiles.
Oono Y, Kawahara Y, Yazawa T, Kanamori H, Kuramata M, Yamagata H, Hosokawa S, Minami H, Ishikawa S, Wu J, Antonio B, Handa H, Itoh T, Matsumoto T. Oono Y, et al. Plant Mol Biol. 2013 Dec;83(6):523-37. doi: 10.1007/s11103-013-0106-4. Epub 2013 Jul 16. Plant Mol Biol. 2013. PMID: 23857470 Free PMC article. - Loss of genetic diversity as a signature of apricot domestication and diffusion into the Mediterranean Basin.
Bourguiba H, Audergon JM, Krichen L, Trifi-Farah N, Mamouni A, Trabelsi S, D'Onofrio C, Asma BM, Santoni S, Khadari B. Bourguiba H, et al. BMC Plant Biol. 2012 Apr 17;12:49. doi: 10.1186/1471-2229-12-49. BMC Plant Biol. 2012. PMID: 22510209 Free PMC article. - Italian weedy rice-A case of de-domestication?
Grimm A, Sahi VP, Amann M, Vidotto F, Fogliatto S, Devos KM, Ferrero A, Nick P. Grimm A, et al. Ecol Evol. 2020 Jul 12;10(15):8449-8464. doi: 10.1002/ece3.6551. eCollection 2020 Aug. Ecol Evol. 2020. PMID: 32788993 Free PMC article. - A chromosome-level genome assembly of the wild rice Oryza rufipogon facilitates tracing the origins of Asian cultivated rice.
Xie X, Du H, Tang H, Tang J, Tan X, Liu W, Li T, Lin Z, Liang C, Liu YG. Xie X, et al. Sci China Life Sci. 2021 Feb;64(2):282-293. doi: 10.1007/s11427-020-1738-x. Epub 2020 Jul 28. Sci China Life Sci. 2021. PMID: 32737856 - Genome-wide patterns of nucleotide polymorphism in domesticated rice.
Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, Bustamante CD, Purugganan MD. Caicedo AL, et al. PLoS Genet. 2007 Sep;3(9):1745-56. doi: 10.1371/journal.pgen.0030163. Epub 2007 Aug 6. PLoS Genet. 2007. PMID: 17907810 Free PMC article.
References
- Oka HI. Origin of cultivated rice. Amsterdam: Elsevier; 1988. Indica-Japonica differentiation of rice cultivars; pp. 141–179.
- Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. - PubMed
- Vitte C, Ishii T, Lamy F, Brar D, Panaud O. Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.) Mol Genet Genomics. 2004;272:504–511. - PubMed
- Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr Opin Plant Mol Biol. 2003;6:139–146. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous