The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression - PubMed (original) (raw)
The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression
Lawrence S Hon et al. Genome Biol. 2007.
Abstract
Background: MicroRNAs (miRNAs) are small noncoding RNAs that bind mRNA target transcripts and repress gene expression. They have been implicated in multiple diseases, such as cancer, but the mechanisms of this involvement are not well understood. Given the complexity and degree of interactions between miRNAs and target genes, understanding how miRNAs achieve their specificity is important to understanding miRNA function and identifying their role in disease.
Results: Here we report factors that influence miRNA regulation by considering the effects of both single and multiple miRNAs targeting human genes. In the case of single miRNA targeting, we developed a metric that integrates miRNA and mRNA expression data to calculate how changes in miRNA expression affect target mRNA expression. Using the metric, our global analysis shows that the repression of a given miRNA on a target mRNA is modulated by 3' untranslated region length, the number of target sites, and the distance between a pair of binding sites. Additionally, we show that some miRNAs preferentially repress transcripts with longer CTG repeats, suggesting a possible role for miRNAs in repeat expansion disorders such as myotonic dystrophy. We also examine the large class of genes targeted by multiple miRNAs and show that specific types of genes are progressively more enriched as the number of targeting miRNAs increases. Expression microarray data further show that these highly targeted genes are downregulated relative to genes targeted by few miRNAs, which suggests that highly targeted genes are tightly regulated and that their dysregulation may lead to disease. In support of this idea, cancer genes are strongly enriched among highly targeted genes.
Conclusion: Our data show that the rules governing miRNA targeting are complex, but that understanding the mechanisms that drive such control can uncover miRNAs' role in disease. Our study suggests that the number and arrangement of miRNA recognition sites can influence the degree and specificity of miRNA-mediated gene repression.
Figures
Figure 1
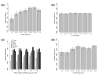
Analysis of the relationship between shorter 3' UTRs and increased repression. The error bars for observed and expected data are based on the distribution of RE values and the distribution of the permutated data, respectively. (a) Shorter 3' UTRs in target genes are more strongly repressed by their predicted cognate miRNAs. (b) The expected RE values (computed using permutation testing) show minimal deviation from 1.0, representing a lack of repression. (c) This trend is increasingly exaggerated when subsets of miRNAs containing larger expression ratios between groups A and B are used, especially in 3' UTRs shorter than 200 bp. (d) The same trend of increased repression in shorter 3' UTRs is observed using a different target prediction algorithm, rna22.
Figure 2
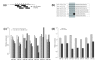
Analysis of site and gene features that affect miRNA repression. The observed values are shown in black; the expected values (computed using permutation testing) are shown in gray. The error bars for observed and expected data are based on the distribution of RE values and the distribution of the permutated data, respectively. (a) Target genes with more binding sites are more strongly repressed. (b) Pairs of binding sites targeted by the same miRNA that are between 16 and 30 bp apart (by start positions) have significantly increased repression (asterisks shown for emphasis). (c) Genes that have multiple pairs of extensively overlapping sites, defined to be two binding sites responsive to the same miRNA whose start positions are within 10 bp of each other, have increased repression.
Figure 3
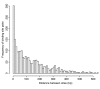
Frequency of pairs of binding sites targeted by the same miRNAs separated by a given distance. The distance between a pair of binding sites is calculated from the 5' ends of the target sites relative to the mRNA. A disproportionate number of binding site pairs are within 10 bp of each other.
Figure 4
CTG repeat-binding miRNAs and their repression of pairs of extensively overlapping sites. (a) A diagram showing how a region of NM_173354 containing seven CTG repeats can result in six binding site seeds (CTGCTG) and five pairs of extensively overlapping sites (pairs of binding sites 3 bp apart). (b) Seven miRNAs containing CAG-rich seed regions that are predicted to bind to CTG repeats. Only hsa-miR-214 has mismatches in the seed region. (c) Number of overlapping binding sites versus relative expression for seven CTG repeat-binding miRNAs. In general, as the number of pairs of extensively overlapping sites increases, the degree of repression increases. In particular, mirs-107, -103, and -15a show a strong correlation. (d) Decreased relative expression of wild-type DMPK with respect to seven CTG repeat-binding miRNAs suggests repression of mutated DMPK by miRNAs could play a role in DM1. Targets with no overlapping pairs of sites served as control and showed no overall repression.
Figure 5
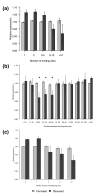
Abundance and functional enrichment of genes targeted by many distinct miRNAs. (a) The observed number of genes targeted by many miRNAs is dramatically greater than the expected number for all three algorithms. The threshold for the number of miRNAs to be considered highly targeted is defined to be one standard deviation more than the average number of miRNAs predicted to target a gene. (b) A large proportion of genes targeted by many miRNAs are transcriptional regulators and nuclear genes, but this enrichment decreases as the number of miRNAs is reduced. Genes involved in ion transporters do not show this trend. In (b-d), asterisks denote P < 0.01. (c) Enrichment, instead of proportion (as before), is shown of transcriptional regulators and nuclear genes for highly targeted genes, with the same enrichment for highly targeted genes. The expected enrichment for a random set of genes targeted by any number of miRNAs is 1.0 (that is, no enrichment), shown by the dotted line. (d) The enrichment of transcriptional regulators and nuclear genes among highly targeted genes remains after controlling for 3' UTR length.
Figure 6
Downregulated expression and enrichment of cancer genes among highly targeted genes. (a) In a comparison of highly targeted genes (n > 20) versus less targeted genes (1 ≤ n ≤ 5) in normal tissue samples [46], 121 out of 158 samples exhibited decreased expression among highly targeted genes (P = 1 × 10-11). (b) Out of 58 NCI60 cancer cell line samples, 58 exhibited decreased expression among highly targeted genes (P = 7 × 10-18). (c) Highly targeted genes are enriched for cancer genes, with cancer genes targeted by >30 miRNAs having the most enrichment. In (c,d), asterisks denote P < 0.01 and crosses denote P < 0.05.(d) This enrichment for cancer genes remains after removing transcriptional regulators, which are prevalent among cancer genes and, as shown earlier, overrepresented among highly targeted genes.
Similar articles
- Regulation of the MIR155 host gene in physiological and pathological processes.
Elton TS, Selemon H, Elton SM, Parinandi NL. Elton TS, et al. Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review. - Unified translation repression mechanism for microRNAs and upstream AUGs.
Ajay SS, Athey BD, Lee I. Ajay SS, et al. BMC Genomics. 2010 Mar 5;11:155. doi: 10.1186/1471-2164-11-155. BMC Genomics. 2010. PMID: 20205738 Free PMC article. - The Emerging Role of MitomiRs in the Pathophysiology of Human Disease.
Duarte FV, Palmeira CM, Rolo AP. Duarte FV, et al. Adv Exp Med Biol. 2015;888:123-54. doi: 10.1007/978-3-319-22671-2_8. Adv Exp Med Biol. 2015. PMID: 26663182 - MicroRNA expression is altered in lateral septum across reproductive stages.
Saul MC, Zhao C, Driessen TM, Eisinger BE, Gammie SC. Saul MC, et al. Neuroscience. 2016 Jan 15;312:130-40. doi: 10.1016/j.neuroscience.2015.11.019. Epub 2015 Nov 17. Neuroscience. 2016. PMID: 26592715 Free PMC article. - microRNA expression in the eyes and their significance in relation to functions.
Xu S. Xu S. Prog Retin Eye Res. 2009 Mar;28(2):87-116. doi: 10.1016/j.preteyeres.2008.11.003. Epub 2008 Nov 28. Prog Retin Eye Res. 2009. PMID: 19071227 Review.
Cited by
- MicroRNAs in metabolism and metabolic disorders.
Rottiers V, Näär AM. Rottiers V, et al. Nat Rev Mol Cell Biol. 2012 Mar 22;13(4):239-50. doi: 10.1038/nrm3313. Nat Rev Mol Cell Biol. 2012. PMID: 22436747 Free PMC article. Review. - Cooperativity between the 3' untranslated region microRNA binding sites is critical for the virulence of eastern equine encephalitis virus.
Trobaugh DW, Sun C, Bhalla N, Gardner CL, Dunn MD, Klimstra WB. Trobaugh DW, et al. PLoS Pathog. 2019 Oct 28;15(10):e1007867. doi: 10.1371/journal.ppat.1007867. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31658290 Free PMC article. - The role of microRNAs in regulating neuronal connectivity.
Chiu H, Alqadah A, Chang C. Chiu H, et al. Front Cell Neurosci. 2014 Jan 3;7:283. doi: 10.3389/fncel.2013.00283. Front Cell Neurosci. 2014. PMID: 24427116 Free PMC article. Review. - Modelling and measuring intracellular competition for finite resources during gene expression.
Sabi R, Tuller T. Sabi R, et al. J R Soc Interface. 2019 May 31;16(154):20180887. doi: 10.1098/rsif.2018.0887. J R Soc Interface. 2019. PMID: 31113334 Free PMC article. - Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins.
Elton TS, Sansom SE, Martin MM. Elton TS, et al. RNA Biol. 2010 Sep-Oct;7(5):540-7. doi: 10.4161/rna.7.5.12685. Epub 2010 Sep 1. RNA Biol. 2010. PMID: 21081842 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources