Switching of the core transcription machinery during myogenesis - PubMed (original) (raw)
Switching of the core transcription machinery during myogenesis
Maria Divina E Deato et al. Genes Dev. 2007.
Abstract
Transcriptional mechanisms that govern cellular differentiation typically include sequence-specific DNA-binding proteins and chromatin-modifying activities. These regulatory factors are assumed necessary and sufficient to drive both divergent programs of proliferation and terminal differentiation. By contrast, potential contributions of the basal transcriptional apparatus to orchestrate cell-specific gene expression have been poorly explored. In order to probe alternative mechanisms that control differentiation, we have assessed the fate of the core promoter recognition complex, TFIID, during skeletal myogenesis. Here we report that differentiation of myoblast to myotubes involves the disruption of the canonical holo-TFIID and replacement by a novel TRF3/TAF3 (TBP-related factor 3/TATA-binding protein-associated factor 3) complex. This required switching of core promoter complexes provides organisms a simple yet effective means to selectively turn on one transcriptional program while silencing many others. Although this drastic but parsimonious transcriptional switch had previously escaped our attention, it may represent a more general mechanism for regulating cell type-specific terminal differentiation.
Figures
Figure 1.
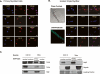
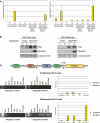
TAF3 and TFIID subunits are differentially expressed in myotubes. (A) Phase-contrast images and DAPI/Phalloidin staining of myoblast, myotubes, and reserve cells from C2C12 cells. (B) Immunoblot analyses of whole-cell extracts from highly purified myoblasts, myotubes, and reserve cells for the expression of TAF3, canonical TFIID subunits, other general transcription factors, and muscle-specific transcription factors and proteins. Ponceau S staining was used to control for equal protein loading per sample. (C–E) Myoblasts, myotubes, and reserve cells were analyzed by immunofluorescence for TAF3, TAF4, TBP, and Pol II localization. Bar, 10 μm.
Figure 2.
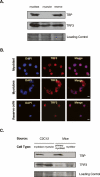
TAF3 and TFIID subunits are differentially expressed in skeletal muscle. (A,B) Primary myoblast cells and myofibers were analyzed by immunofluorescence for TAF3, TAF4, TBP, and Pol II staining patterns. Also included are the phase-contrast image and DAPI/Phalloidin staining of the isolated myofibers. Bar, 10 μm. (C) Immunoblot analyses of TAF3, TAF4, TBP, and RNA Pol II protein levels in primary myoblast cells and myofibers. As controls, the expression of these factors was compared with their expression profiles in C2C12 myoblast and myotube cells. Myf5 and Myosin expression in primary myoblast cells or myofibers were also analyzed as additional controls. Ponceau S staining was used as loading control.
Figure 3.
TRF3 Expression in terminally differentiated myotubes and myofibers. (A) Immunoblot analyses for TRF3 expression in myoblasts, myotubes, and reserve cell populations. (B) Representative cell immunofluorescence analyses of TRF3 localization in various C2C12 cell types. Bar, 10 μm. (C) Immunoblot analyses of TRF3 expression in isolated myofibers. Ponceau S was used to assess the total protein loaded per sample for immunoblot analyses.
Figure 4.
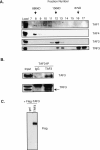
TAF3 and TRF3 form a complex in differentiated myotubes. (A) Size exclusion chromatography of nuclear extracts prepared from differentiated C2C12 cells that contain both myotubes and reserve cell populations. Fractions were analyzed by immunoblot for TAF3 and TRF3. The representative molecular weights of protein standards are indicated in kilodaltons. (B) TAF3-specific antibody was used to coimmunoprecipitate associated proteins from myotube nuclear extracts and was subsequently analyzed by immunoblot. (C) In vitro binding assay using Glutathione Sepharose-bound gst or gst-TRF3- and Flag-TAF3-transfected SF9 protein extracts. Samples were analyzed by immunoblot.
Figure 5.
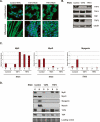
TAF3 and TRF3 are required factors for myoblast differentiation. (A) Representative images of DAPI/Phalloidin staining using control, TAF3, and TRF3 RNAi cell lines grown under proliferation and differentiation conditions. (B) TAF3 and TRF3 RNAi myoblast cells were analyzed by immunoblot for TAF3, TAF4, TBP, and TRF3 protein levels. Anti-tubulin antibody was used as loading control. (C,D) Control, TAF3, and TRF3 RNAi cells grown under either proliferation (P) or differentiation (D) conditions, were analyzed by Q-PCR and immunoblot for Myf5, MyoD, and Myogenin levels. Each sample was normalized to the abundance of U6 RNA and presented as means ± SD from four independent reactions. In addition, the protein levels of TAF4 and TBP were also analyzed. Ponceau S stain was used to control for equal protein loading.
Figure 6.
TAF3 and TRF3 are enriched on the myogenin promoter in differentiated C2C12 cells. (A) The rescue phenotype was analyzed by Q-PCR for the recovery of Myogenin expression in Flag-TAF3- or Flag-TRF3-rescued TAF3-RNAi or TRF3-RNAi cell lines. Samples were normalized to U6 RNA levels, and are presented as mean ± SD from four independent reactions. (B) The rescued RNAi cell lines grown in either proliferation (P) or differentiation (D) conditions were analyzed for endogenous levels of Myogenin by immunoblot. The expression levels of the tagged versions of TAF3 and TRF3 used in the rescue experiment were also analyzed. Ponceau S stain was used as loading control. (C) The mouse Myogenin promoter consists of a TATA box and is flanked by activator sites (MEF2 and E-box). Arrowheads correspond to the region spanning the Myogenin primer sequences used to analyze the ChIP. Intergenic primers were used to amplify a region ∼10 kb upstream of the Myogenin transcription start site. ChIP from either proliferating or differentiated C2C12 cells (after 3 d of induction) using the indicated antibodies. ChIP samples were also analyzed by Q-PCR using primers directed at either intergenic regions or the Myogenin core promoter region. Values were normalized to input and presented as percent of input. The data reflect the mean ± SD from four independent reactions.
Figure 7.
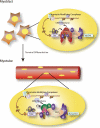
Proposed model for transcription initiation in myoblast and myotube cells. The novel core transcription initiation complex composed of TAF3 and TRF3 functionally replaces the canonical TFIID complex during the differentiation of myoblasts into myotubes. This mechanism of switching the core transcription machinery may regulate the unique transcriptional program during cell type-specific terminal differentiation.
Comment in
- Transcription strategies in terminally differentiated cells: shaken to the core.
Jones KA. Jones KA. Genes Dev. 2007 Sep 1;21(17):2113-7. doi: 10.1101/gad.1598007. Genes Dev. 2007. PMID: 17785521 No abstract available.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.
Love AMA, Edwards C, Cai RY, Gibbs V. Love AMA, et al. Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - "I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.
Huang Y, Trollor JN, Foley KR, Arnold SRC. Huang Y, et al. Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article. - Harnessing Biological Insight to Accelerate Tuberculosis Drug Discovery.
de Wet TJ, Warner DF, Mizrahi V. de Wet TJ, et al. Acc Chem Res. 2019 Aug 20;52(8):2340-2348. doi: 10.1021/acs.accounts.9b00275. Epub 2019 Jul 30. Acc Chem Res. 2019. PMID: 31361123 Free PMC article. Review.
Cited by
- Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7.
Kloet SL, Whiting JL, Gafken P, Ranish J, Wang EH. Kloet SL, et al. Mol Cell Biol. 2012 Aug;32(16):3358-69. doi: 10.1128/MCB.00416-12. Epub 2012 Jun 18. Mol Cell Biol. 2012. PMID: 22711989 Free PMC article. - Large Polyglutamine Repeats Cause Muscle Degeneration in SCA17 Mice.
Huang S, Yang S, Guo J, Yan S, Gaertig MA, Li S, Li XJ. Huang S, et al. Cell Rep. 2015 Oct 6;13(1):196-208. doi: 10.1016/j.celrep.2015.08.060. Epub 2015 Sep 17. Cell Rep. 2015. PMID: 26387956 Free PMC article. - Identification of myelin transcription factor 1 (MyT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex.
Yokoyama A, Igarashi K, Sato T, Takagi K, Otsuka I M, Shishido Y, Baba T, Ito R, Kanno J, Ohkawa Y, Morohashi K, Sugawara A. Yokoyama A, et al. J Biol Chem. 2014 Jun 27;289(26):18152-62. doi: 10.1074/jbc.M114.566448. Epub 2014 May 14. J Biol Chem. 2014. PMID: 24828497 Free PMC article. - H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation.
Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. Lauberth SM, et al. Cell. 2013 Feb 28;152(5):1021-36. doi: 10.1016/j.cell.2013.01.052. Cell. 2013. PMID: 23452851 Free PMC article. - Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression.
Faralli H, Dilworth FJ. Faralli H, et al. Comp Funct Genomics. 2012;2012:836374. doi: 10.1155/2012/836374. Epub 2012 Jun 28. Comp Funct Genomics. 2012. PMID: 22811619 Free PMC article.
References
- Albright S.R., Tjian R., Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. - PubMed
- Bartfai R., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Hadzhiev Y., Tora L., Orban L., Muller F., Tora L., Orban L., Muller F., Orban L., Muller F., Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 2004;14:593–598. - PubMed
- Berkes C.A., Tapscott S.J., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. - PubMed
- Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Sharan R., Kluger Y., Dynlacht B.D., Kluger Y., Dynlacht B.D., Dynlacht B.D. An initial blueprint for myogenic differentiation. Genes & Dev. 2005;19:553–569. - PMC - PubMed
- Brunkhorst A., Karlen M., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Karlen M., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Nelson M.A., Metsis M., Hermanson O., Metsis M., Hermanson O., Hermanson O. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol. Cell. Neurosci. 2005;29:250–258. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases