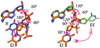Enzymatic capture of an extrahelical thymine in the search for uracil in DNA - PubMed (original) (raw)
. 2007 Sep 27;449(7161):433-7.
doi: 10.1038/nature06131. Epub 2007 Aug 19.
Affiliations
- PMID: 17704764
- PMCID: PMC2754044
- DOI: 10.1038/nature06131
Enzymatic capture of an extrahelical thymine in the search for uracil in DNA
Jared B Parker et al. Nature. 2007.
Abstract
The enzyme uracil DNA glycosylase (UNG) excises unwanted uracil bases in the genome using an extrahelical base recognition mechanism. Efficient removal of uracil is essential for prevention of C-to-T transition mutations arising from cytosine deamination, cytotoxic U*A pairs arising from incorporation of dUTP in DNA, and for increasing immunoglobulin gene diversity during the acquired immune response. A central event in all of these UNG-mediated processes is the singling out of rare U*A or U*G base pairs in a background of approximately 10(9) T*A or C*G base pairs in the human genome. Here we establish for the human and Escherichia coli enzymes that discrimination of thymine and uracil is initiated by thermally induced opening of T*A and U*A base pairs and not by active participation of the enzyme. Thus, base-pair dynamics has a critical role in the genome-wide search for uracil, and may be involved in initial damage recognition by other DNA repair glycosylases.
Figures
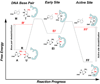
Figure 1. Extrahelical uracil recognition by UNG and reaction coordinate tuning
Uracil emerges from the DNA base stack (reactant or R state) by spontaneous U:A base pair breathing, where it is then trapped by UNG as an unstable early intermediate state (EI) on the base flipping reaction coordinate. EI is very unstable (high energy), compared to the low energy intrahelical bound state for a T:A or U:A base pair or the fully-flipped (FF) state , . Substitution of adenine with its nonpolar analogue, 4-methylindole (M), energetically destabilizes the intrahelical R′ state. Substitution of uracil with 5-methyluracil (5-MeU or thymine, T) greatly destabilizes the FF′ state because the bulkier T fits poorly into the uracil active site, but T can access the EI′ state , . The energetic effects of reaction coordinate tuning on base flipping are shown by the vertical arrows. Destabilization of the R and FF states allows population of the otherwise unstable EI intermediate, allowing its structural characterization by X-ray crystallography. The free energy levels that are depicted in this Figure are exaggerated for clarity of exposition.
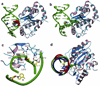
Figure 2. Stabilization of extrahelical thymine in the EI′ state in complex with UNG
(A) View of the extrahelical thymine trapped in the EI′ state with UNG. (B) View of extrahelical uracil containing DNA in the FF state with UNG (pdb code 1EMH) . (C) Interactions with the MT base pair in the EI′ state. (D) Computational model of the R complex with an intrahelical thymine (blue) is shown aligned with the crystallographic models for the EI’ (red) and FF (green) complexes. The view is rotated ninety degrees out of the plane of the page relative to panels (A) and (B). The extended coil region containing His148 is indicated in red.
Figure 3. Conformational changes in the sugar and base along the flipping reaction coordinate
In the first step of the reaction to form EI, the sugar plane rotates about an apparent angle of 30°, and the base rotates 180° around the glycosidic bond and moves about 4.4 Å relative to the B DNA reactant state (R, blue). In the second step (EI➔FF), the sugar plane and base rotate a further 120° and 90°, respectively. Note that the structure of the FF state was obtained using the C-glycoside analogue of deoxyuridine, pseudodeoxyuridine. Changes in the DNA backbone torsional angles that accompany these transformations are listed in the figure.
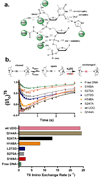
Figure 4. Interaction map for the EI′ complex and imino proton exchange profiles for wtUNG and several mutant forms
(A) Side chain and backbone interactions of UNG with the extrahelical thymine base and DNA phosphate backbone in the EI′ complex. (B) Imino proton exchange is a two step process that requires base pair opening (_K_op = _k_open/_k_close) followed by imino proton exchange with solvent which may be followed by using NMR magnetization transfer from water . The data are fitted to eq 1 and the exchange time courses for wtUNG and six mutants are indicated. (C) Exchange rates (_k_ex) for T6 in the presence of wtUNG and the indicated mutants.
Similar articles
- The catalytic power of uracil DNA glycosylase in the opening of thymine base pairs.
Cao C, Jiang YL, Krosky DJ, Stivers JT. Cao C, et al. J Am Chem Soc. 2006 Oct 11;128(40):13034-5. doi: 10.1021/ja062978n. J Am Chem Soc. 2006. PMID: 17017766 Free PMC article. - Dynamics of uracil and 5-fluorouracil in DNA.
Parker JB, Stivers JT. Parker JB, et al. Biochemistry. 2011 Feb 8;50(5):612-7. doi: 10.1021/bi101536k. Epub 2011 Jan 13. Biochemistry. 2011. PMID: 21190322 Free PMC article. - Structure and function in the uracil-DNA glycosylase superfamily.
Pearl LH. Pearl LH. Mutat Res. 2000 Aug 30;460(3-4):165-81. doi: 10.1016/s0921-8777(00)00025-2. Mutat Res. 2000. PMID: 10946227 Review. - Dynamic opening of DNA during the enzymatic search for a damaged base.
Cao C, Jiang YL, Stivers JT, Song F. Cao C, et al. Nat Struct Mol Biol. 2004 Dec;11(12):1230-6. doi: 10.1038/nsmb864. Epub 2004 Nov 21. Nat Struct Mol Biol. 2004. PMID: 15558051 - Properties and functions of human uracil-DNA glycosylase from the UNG gene.
Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Krokan HE, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:365-86. doi: 10.1016/s0079-6603(01)68112-1. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554311 Review.
Cited by
- Inhibition of Human Uracil DNA Glycosylase Sensitizes a Large Fraction of Colorectal Cancer Cells to 5-Fluorodeoxyuridine and Raltitrexed but Not Fluorouracil.
Christenson ES, Gizzi A, Cui J, Egleston M, Seamon KJ, DePasquale M, Orris B, Park BH, Stivers JT. Christenson ES, et al. Mol Pharmacol. 2021 Jun;99(6):412-425. doi: 10.1124/molpharm.120.000191. Epub 2021 Apr 1. Mol Pharmacol. 2021. PMID: 33795350 Free PMC article. - The cis-(5R,6S)-thymine glycol lesion occupies the wobble position when mismatched with deoxyguanosine in DNA.
Brown KL, Basu AK, Stone MP. Brown KL, et al. Biochemistry. 2009 Oct 20;48(41):9722-33. doi: 10.1021/bi900695e. Biochemistry. 2009. PMID: 19772348 Free PMC article. - Characterization of DNA with an 8-oxoguanine modification.
Singh SK, Szulik MW, Ganguly M, Khutsishvili I, Stone MP, Marky LA, Gold B. Singh SK, et al. Nucleic Acids Res. 2011 Aug;39(15):6789-801. doi: 10.1093/nar/gkr275. Epub 2011 May 13. Nucleic Acids Res. 2011. PMID: 21572101 Free PMC article. - A two-step nucleotide-flipping mechanism enables kinetic discrimination of DNA lesions by AGT.
Hu J, Ma A, Dinner AR. Hu J, et al. Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4615-20. doi: 10.1073/pnas.0708058105. Epub 2008 Mar 19. Proc Natl Acad Sci U S A. 2008. PMID: 18353991 Free PMC article. - A perspective on the molecular simulation of DNA from structural and functional aspects.
Mondal M, Yang L, Cai Z, Patra P, Gao YQ. Mondal M, et al. Chem Sci. 2021 Mar 15;12(15):5390-5409. doi: 10.1039/d0sc05329e. Chem Sci. 2021. PMID: 34168783 Free PMC article. Review.
References
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. - PubMed
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA Repair Glycosylases. Chem. Rev. 2003;103:2729–2759. - PubMed
- Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA-General mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2006 - PubMed
- Slupphaug G, et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA [see comments] Nature. 1996;384:87–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases