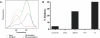Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3 - PubMed (original) (raw)
Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3
Kimberly J Reese et al. PLoS Genet. 2007 Aug.
Abstract
Paternal repression of the imprinted H19 gene is mediated by a differentially methylated domain (DMD) that is essential to imprinting of both H19 and the linked and oppositely imprinted Igf2 gene. The mechanisms by which paternal-specific methylation of the DMD survive the period of genome-wide demethylation in the early embryo and are subsequently used to govern imprinted expression are not known. Methyl-CpG binding (MBD) proteins are likely candidates to explain how these DMDs are recognized to silence the locus, because they preferentially bind methylated DNA and recruit repression complexes with histone deacetylase activity. MBD RNA and protein are found in preimplantation embryos, and chromatin immunoprecipitation shows that MBD3 is bound to the H19 DMD. To test a role for MBDs in imprinting, two independent RNAi-based strategies were used to deplete MBD3 in early mouse embryos, with the same results. In RNAi-treated blastocysts, paternal H19 expression was activated, supporting the hypothesis that MBD3, which is also a member of the Mi-2/NuRD complex, is required to repress the paternal H19 allele. RNAi-treated blastocysts also have reduced levels of the Mi-2/NuRD complex protein MTA-2, which suggests a role for the Mi-2/NuRD repressive complex in paternal-specific silencing at the H19 locus. Furthermore, DNA methylation was reduced at the H19 DMD when MBD3 protein was depleted. In contrast, expression and DNA methylation were not disrupted in preimplantation embryos for other imprinted genes. These results demonstrate new roles for MBD3 in maintaining imprinting control region DNA methylation and silencing the paternal H19 allele. Finally, MBD3-depleted preimplantation embryos have reduced cell numbers, suggesting a role for MBD3 in cell division.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
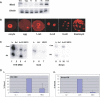
Figure 1. Mbd3 RNA and Protein Patterns in Oocytes and Preimplantation Embryos
(A) Mbd3 and globin primers were used to amplify Mbd3 and exogenously added globin RNA from oocyte and preimplantation embryo cDNA samples. Mbd3 RNA was normalized against exogenously added globin RNA. (B) Immunocytochemistry using an anti-MBD3 antibody and a Cy3 secondary antibody showed that MBD3, a nuclear protein, is also maternally contributed to oocytes and follows a pattern similar to that of Mbd3 RNA, increasing to high levels at the blastocyst stage. (C) Chromatin immunoprecipitation (ChIP) from hybrid ES cells at the H19 DMD, Oct4, and Snrpn loci with acetylated H3 antibody (AcH3) (Lanes 3 and 10) and MBD3 antibody (Lanes 4, 7, and 11). The H19 DMD and Oct4 are precipitated with the MBD3 antibody. Input chromatin (Lanes 1, 5, and 8) and no primary antibody (Lanes 2, 6, and 9) are included as positive and negative PCR controls. In the experiment presented here, 65% of the chromatin associated with the MBD3 antibody was from the paternal allele (C). (D) Quantitative real-time PCR was conducted using primers specific to the H19 DMD (i) or to the Snrpn imprinting control region (ii) for antibody-precipitated samples and no primary antibody (No 1) controls. The average percentage (+/− standard deviation) of bound material for three or more chromatin preparations is shown. The AcH3 results were set at 100% and the other values were normalized to the AcH3 results. A significant enrichment of MBD3 at the H19 DMD was evident when compared with the no-primary control (p < 0.05), but this enrichment was not observed at the Snrpn imprinting control region (p = 0.12). There was a significant enrichment for AcH3 histones at both loci (p < 0.01).
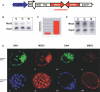
Figure 2. Mbd3 RNA and Protein Are Specifically Reduced in RNAi-Treated Embryos
(A) A schematic depicting the RNAi transgene showing the Zp3 promoter driving expression of Gfp and the Mbd3 inverted repeat in growing oocytes. The dsRNA used in the injection RNAi experiments is designated by the red line below the inverted Mbd3 repeat and is identical to the sequence used in the TG RNAi. (B) RNA was collected from embryos injected with dsMbd3 [M], dsGfp [G], or uninjected [U], cultured for 96 h, and collected in pools of six blastocyst-stage embryos. The RNA was used for quantitative RT-PCR. When normalized to Gapd RNA levels and compared to uninjected and dsGfp-injected control embryos, dsMbd3-injected embryos show reduced levels of RNA. The embryos shown have only 24% of the Mbd3 RNA compared to uninjected embryos. RNA from TG and NTG GV stage oocytes was also used for quantitative RT-PCR. (C) A graphical representation of real-time quantitative RT-PCR experiments demonstrating reduced Mbd3 RNA levels in TG oocytes. TG and NTG bars represent the ratios of Mbd3 to H2afy2 levels (a H2 histone expressed in early embryos) derived from crossing points. The ratios are normalized to the levels in NTG embryos (shown as 1), and are the averages of three experiments. TG oocytes have only 15% of the Mbd3 RNA compared to NTG oocytes. (D) RT-PCR analysis for Mbd2 demonstrates targeting specificity for Mbd3 because no reduction in Mbd2 mRNA was seen. (E) Embryos were subjected to immunocytochemistry with the anti-MBD3 antibody as described in Figure 1B. MBD3 protein levels are greatly reduced in both dsMbd3 injected embryos (iv) and TG embryos (viii) when compared to dsGfp injected (ii) and NTG (vi) controls. Embryos were also treated with SytoX (i, iii) or DAPI (v, vii) to show the nuclei of the embryos. DsGfp injected embryos are shown in i, ii; dsMbd3 injected embryos are shown in iii, iv; with NTG (v, vi) and TG blastocysts (vii, viii) shown in the bottom panels.
Figure 3. H19 Is Derepressed at the Paternal Allele after Mbd3 RNAi
One-cell C57BL/6J X B6(CAST7) embryos were injected with dsRNA, cultured to the blastocyst stage, and collected singly for RNA extraction. Similarly, TG and NTG embryos were either cultured as above or collected from mothers at 3.5 dpc. Real-time RT-PCR with allele-specific hybridization probes was used to determine H19 expression from each allele, as shown in (A). Shown are the melting curves of the control samples along with a biallelic TG and monoallelic TG sample (the melting curves for the ten other samples processed at the same time are omitted for clarity). The red curve is the melting curve of a Cast control (probe melts off at 60 °C) and the green represents a C57BL/6J control (probe melts off at 65 °C). The grey curve is one of the biallelic TG blastocysts, with two peaks corresponding to the paternal Cast allele and the maternal C57BL/6J allele. The blue curve is a TG sample that is monoallelic for H19 (maternal C57BL/6J allele only). (B) H19 is biallelically expressed in 26% (9/35) of dsMbd3 injected embryos. This is significantly different from controls (p < 0.01 compared to uninjected [2/46] and p < 0.01 compared to dsGfp injected [0/28]). Most control embryos (uninjected and dsGfp) express only the maternal H19 allele. In the Zp3-dsMbd3 TG embryos, 40% (15/38) of TG embryos show biallelic expression of H19. None (0/5) of the NTG controls showed biallelic expression.
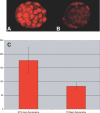
Figure 4. The Mi-2/NuRD Complex Protein MTA2 Is Reduced in Mbd3 RNAi Blastocysts
(A, B) NTG (n = 4) (A) and Mbd3 RNAi TG (n = 16) (B) embryos were subjected to immunocytochemistry with the anti-MTA2 antibody. When Mbd3 is reduced in TG embryos, MTA2 protein is significantly reduced (compare [A] to [B]). (C) Measurement of mean nuclear fluorescence using ImageJ software of confocal images shows that the mean fluorescence of TG embryos have on average 47% of the nuclear fluorescent MTA2 signal compared to NTG samples.
Figure 5. Reduction of Mbd3 in Embryos Results in a Loss of Methylation at the H19 DMD
One-cell C57BL/6J X B6(CAST7) embryos were injected with dsRNA, cultured to the blastocyst stage, and collected in pools of five blastocysts (top). Similarly, TG and NTG embryos were isolated at the one-cell stage, cultured to the blastocyst stage, and collected in pools of five (bottom). Bisulfite mutagenesis was performed on agarose-embedded pools of five blastocysts. CpGs (17) were assayed in the 5′ half of the DMD as indicated by the black line in the diagram in the center. Each line of circles represents a single DNA strand with the number to the left of the line corresponding to the number of times this pattern was seen. Each circle represents a single CpG. If the CpG was methylated, the circle is filled. Those strands with less than half of the CpGs methylated are considered hypomethylated. In control embryos, only a few hypomethylated strands are recovered (13% and 17% for uninjected and dsGFP-injected blastocysts, respectively), but in dsMbd3-injected embryos, 42% of the paternal strands are hypomethylated. TG RNAi blastocysts showed similar results with 42% of the paternal strands hypomethylated compared to 16% of the strands in NTG embryos. Only paternal strands are shown, as all sequenced maternal strands were unmethylated as expected. Although its expression also depends on the DMD, Igf2 is not expressed at this stage.
Similar articles
- Metastasis tumor antigen 2 (MTA2) is involved in proper imprinted expression of H19 and Peg3 during mouse preimplantation development.
Ma P, Lin S, Bartolomei MS, Schultz RM. Ma P, et al. Biol Reprod. 2010 Dec;83(6):1027-35. doi: 10.1095/biolreprod.110.086397. Epub 2010 Aug 18. Biol Reprod. 2010. PMID: 20720167 Free PMC article. - Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus.
Thorvaldsen JL, Fedoriw AM, Nguyen S, Bartolomei MS. Thorvaldsen JL, et al. Mol Cell Biol. 2006 Feb;26(4):1245-58. doi: 10.1128/MCB.26.4.1245-1258.2006. Mol Cell Biol. 2006. PMID: 16449639 Free PMC article. - The loss of imprinted DNA methylation in mouse blastocysts is inflicted to a similar extent by in vitro follicle culture and ovulation induction.
Saenz-de-Juano MD, Billooye K, Smitz J, Anckaert E. Saenz-de-Juano MD, et al. Mol Hum Reprod. 2016 Jun;22(6):427-41. doi: 10.1093/molehr/gaw013. Epub 2016 Feb 7. Mol Hum Reprod. 2016. PMID: 26908643 - Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation.
Sasaki H, Ishihara K, Kato R. Sasaki H, et al. J Biochem. 2000 May;127(5):711-5. doi: 10.1093/oxfordjournals.jbchem.a022661. J Biochem. 2000. PMID: 10788777 Review.
Cited by
- Genetic Studies on Mammalian DNA Methyltransferases.
Dan J, Chen T. Dan J, et al. Adv Exp Med Biol. 2022;1389:111-136. doi: 10.1007/978-3-031-11454-0_5. Adv Exp Med Biol. 2022. PMID: 36350508 Free PMC article. - Genomic Imprinting in the New Omics Era: A Model for Systems-Level Approaches.
Hubert JN, Demars J. Hubert JN, et al. Front Genet. 2022 Mar 15;13:838534. doi: 10.3389/fgene.2022.838534. eCollection 2022. Front Genet. 2022. PMID: 35368671 Free PMC article. Review. - Stability and Lability of Parental Methylation Imprints in Development and Disease.
Farhadova S, Gomez-Velazquez M, Feil R. Farhadova S, et al. Genes (Basel). 2019 Dec 2;10(12):999. doi: 10.3390/genes10120999. Genes (Basel). 2019. PMID: 31810366 Free PMC article. Review. - H19 gene methylation status is associated with male infertility.
Li XP, Hao CL, Wang Q, Yi XM, Jiang ZS. Li XP, et al. Exp Ther Med. 2016 Jul;12(1):451-456. doi: 10.3892/etm.2016.3314. Epub 2016 May 9. Exp Ther Med. 2016. PMID: 27347077 Free PMC article. - Promyelocytic leukemia zinc finger-retinoic acid receptor α (PLZF-RARα), an oncogenic transcriptional repressor of cyclin-dependent kinase inhibitor 1A (p21WAF/CDKN1A) and tumor protein p53 (TP53) genes.
Choi WI, Yoon JH, Kim MY, Koh DI, Licht JD, Kim K, Hur MW. Choi WI, et al. J Biol Chem. 2014 Jul 4;289(27):18641-56. doi: 10.1074/jbc.M113.538777. Epub 2014 May 12. J Biol Chem. 2014. PMID: 24821728 Free PMC article.
References
- O'Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum Mol Genet. 2005;14(Spec No 1):R113–R120. - PubMed
- Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–259. - PubMed
- Weksberg R, Shuman C, Smith AC. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137:12–23. - PubMed
- Reese KJ, Bartolomei MS. Establishment and maintenance of H19 imprinting in the germline and preimplantation embryo. Cytogenet Genome Res. 2006;113:153–158. - PubMed
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous