The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes - PubMed (original) (raw)
The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes
Charlotte Barrandon et al. Mol Cell Biol. 2007 Oct.
Abstract
The positive transcription elongation factor P-TEFb controls the elongation of transcription by RNA polymerase II. P-TEFb is inactivated upon binding to HEXIM1 or HEXIM2 proteins associated with a noncoding RNA, 7SK. In response to the inhibition of transcription, 7SK RNA, as well as HEXIM proteins, is released by an unknown mechanism and P-TEFb is activated. New partners of 7SK RNA were searched for as potential players in this feedback process. A subset of heterogeneous ribonuclear proteins, hnRNPs Q and R and hnRNPs A1 and A2, were thus identified as major 7SK RNA-associated proteins. The degree of association of 7SK RNA with these hnRNPs increased when P-TEFb-HEXIM1-7SK was dissociated following the inhibition of transcription or HEXIM1 knockdown. This finding suggested that 7SK RNA shuttles from HEXIM1-P-TEFb complexes to hnRNPs. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK complexes was attenuated when both hnRNPs A1 and A2 were knocked down by small interfering RNA. As hnRNPs are known to interact transiently with RNA while it is synthesized, hnRNPs released from nascent transcripts may trap 7SK RNA and thereby contribute to the activation of P-TEFb.
Figures
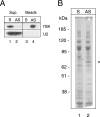
FIG. 1.
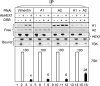
Purification of 7SK-associated proteins. Glycerol gradient fractions containing 7SK RNA were incubated with 7SK sense (S) or antisense (AS) biotinylated 2′-O-methylated oligoribonucleotides and streptavidin beads. Beads and supernatants (Sup.) were separated by centrifugation. (A) 7SK or U2 RNAs were detected by Northern blotting. (B) Proteins bound to streptavidin beads were stained by Coomassie blue after elution and SDS-polyacrylamide gel electrophoresis. The star indicates the protein identified as an hnRNP Q form by mass spectrometry.
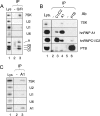
FIG. 2.
Immunoprecipitation of 7SK RNA with hnRNP Q and R and hnRNP A1 proteins. Proteins and RNAs were immunoprecipitated with anti-hnRNP Q/R (A); anti-hnRNP C1/C2, anti-hnRNP A1, and anti-PTB antibodies (Ab) (B); and anti-hnRNP A1 antibody (C). Mock antibodies (-) were used for lanes 2 in panels A and C and lanes 2 and 5 in panel B. Proteins in input lysates (Lys) and protein A beads were detected by Western blotting. RNAs were probed by Northern blotting using the same samples. IP, immunoprecipitate.
FIG. 3.
7SK RNA binding involves RRMs. Asynchronously growing cells were transiently transfected to express tagged hnRNPs or empty vector plasmids (-). Transfected cells were exposed to DRB (100 μM; +) or not (−) for 1 h prior to lysis. (A) RNAs immunoprecipitated with full-length (wild-type [WT]) or truncated (ΔRRM and ΔSMN) HA-tagged hnRNP R proteins. IP, immunoprecipitate. (B) RNAs were retained on biotin beads with wild-type TAP-tagged hnRNP A1 proteins or mutant forms carrying F→D (F>D) point mutations in either RRM1 or RRM2. HA-tagged hnRNP R or TAP-tagged hnRNP A1 on beads was detected by Western blotting with anti-HA or anti-hnRNP A1, respectively. RNAs were probed by Northern blotting using the same samples. Samples in all lanes were prepared from the same number of transfected cells. (C) The mobility of synthetic 7SK RNA decreased upon the addition of increasing amounts of GST-hnRNP A1 (lanes 5 to 7). The addition of genuine GST had no effect (lanes 2 to 4).
FIG. 4.
Association of 7SK with hnRNP A1 increases upon P-TEFb-HEXIM1-7SK dissociation. Cell lysates were subjected to immunoprecipitation (IP) with antibodies against HEXIM1 (HEX1), cyclin T1 (T1), hnRNP A1 (A1), or preimmune serum (mock). Histograms correspond to the quantification of immunoprecipitated (bound) 7SK by Northern blotting. Lysates were probed by Western blotting for cyclin T1, hnRNP A1, HEXIM1 (HEX1), HEXIM2 (HEX2), and actin (Act). (A) Cycloheximide-treated cells were treated with DRB (100 μM; + and ±) or not (−) for 1 h. DRB-treated cells were (±) or were not (+) allowed to recover in normal medium for 1 h prior to lysis. Cycloheximide (50 μg ml−1) was present throughout the entire experiment. (B) Cells were treated with actinomycin D (Act D) at 1 μg ml−1 (+) or not (−) for 1 h. (C) Cells were treated for 48 h with interfering RNA (RNAi) targeting vimentin (Vim) or HEXIM1 (Hex).
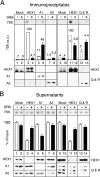
FIG. 5.
Association of 7SK with hnRNPs A1 and A2 and hnRNPs Q and R increases in asynchronously growing cells exposed to DRB. HeLa cells were treated with DRB (100 μM; +) or not (−) for 1 h. Lysates were incubated with either mock serum or anti-HEXIM1, anti-hnRNP A1, anti-hnRNP A2, or anti-hnRNP Q/R. Immunoprecipitates (A) and postimmunoprecipitation supernatants (B) were probed for 7SK RNA and proteins by Northern or Western blotting. Histograms correspond to 7SK quantification by Northern blotting. In panel A, the amount of 7SK RNA immunoprecipitated with HEXIM1 (HEX1) from untreated cell lysates was set at 100 a.u.
FIG. 6.
Complete extraction of hnRNPs A1 and A2 and hnRNPs Q and R from mitotic cells. Lysates from asynchronous (A) or mitotic (B) cells treated with DRB (100 μM; +) or not (−) and treated with RNase (10 μg ml−1; +) or not (−) were fractionated by centrifugation at 10,000 × g into supernatants (Sup) and nuclear pellets (N. Pel). RNA polymerase II (Rpb1) and hnRNPs were detected by Western blotting. 7SK RNA was detected by Northern blotting.
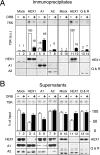
FIG. 7.
7SK RNA transfer from HEXIM1 to hnRNPs Q and R or to hnRNPs A1 and A2 in DRB-treated mitotic cells. HeLa cells arrested in mitosis were collected after being treated for 4 h with nocodazole in the absence (−) or in the presence (+) of DRB (100 μM). Lysates were incubated with either mock serum or anti-HEXIM1, anti-hnRNP A1, anti-hnRNP A2, or anti-hnRNP Q/R. Immunoprecipitates (A) and postimmunoprecipitation supernatants (B) were probed for 7SK RNA and proteins by Northern or Western blotting. Histograms correspond to 7SK quantification by Northern blotting. In panel A, the amount of 7SK RNA immunoprecipitated with HEXIM1 (HEX1) from untreated cell lysates was set at 100 a.u.
FIG. 8.
Decreased DRB-induced dissociation of P-TEFb-HEXIM1-7SK RNA following RNAi. HeLa cells were treated with interfering RNA targeting vimentin or hnRNPs A1 and A2 and exposed to DRB (+) or not (−) for 1 h prior to lysis. Lysates were subjected to immunoprecipitation (IP) with anti-HEXIM1 (AbHEX1; +) or mock (−) antibodies. The presence of hnRNPs A1 and A2 and HEXIM1 (HEX1) in supernatants (free) or on protein A beads (bound) was tested by Western blotting. 7SK RNA immunoprecipitated with HEXIM1 was detected by Northern blotting. Signal quantification (the average of results from three independent experiments) is shown in the lower panel. Histograms correspond to the quantification of immunoprecipitated (bound) 7SK by Northern blotting. The level of 7SK RNA immunoprecipitated with HEXIM1 from cells that had not been exposed to DRB was set at 100 a.u.
FIG. 9.
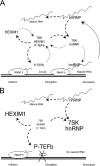
Model of coupled hnRNP-7SK and P-TEFb-HEXIM1-7SK RNA complex formation. (A) At the initiation of transcription, P-TEFb is loaded onto the transcription machinery and 7SK RNA is released from P-TEFb (and HEXIM1) and transferred onto an hnRNP that has been released from a mature RNA molecule (dashed-line arrows). Upon the elongation or termination of transcription, as hnRNPs are recruited onto the nascent transcripts, they release their 7SK RNA companion, which reassembles onto P-TEFb (dotted-line arrows). RNAP II, RNA polymerase II. (B) There are no nascent chains when transcription is inhibited. Core P-TEFb accumulates at the expense of P-TEFb-HEXIM-7SK. hnRNPs are released and trap 7SK RNA.
Similar articles
- Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb.
Van Herreweghe E, Egloff S, Goiffon I, Jády BE, Froment C, Monsarrat B, Kiss T. Van Herreweghe E, et al. EMBO J. 2007 Aug 8;26(15):3570-80. doi: 10.1038/sj.emboj.7601783. Epub 2007 Jul 5. EMBO J. 2007. PMID: 17611602 Free PMC article. - Hexim1 sequesters positive transcription elongation factor b from the class II transactivator on MHC class II promoters.
Kohoutek J, Blazek D, Peterlin BM. Kohoutek J, et al. Proc Natl Acad Sci U S A. 2006 Nov 14;103(46):17349-54. doi: 10.1073/pnas.0603079103. Epub 2006 Nov 6. Proc Natl Acad Sci U S A. 2006. PMID: 17088550 Free PMC article. - siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes.
Haaland RE, Herrmann CH, Rice AP. Haaland RE, et al. J Cell Physiol. 2005 Dec;205(3):463-70. doi: 10.1002/jcp.20528. J Cell Physiol. 2005. PMID: 16152622 - 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor.
Diribarne G, Bensaude O. Diribarne G, et al. RNA Biol. 2009 Apr-Jun;6(2):122-8. doi: 10.4161/rna.6.2.8115. Epub 2009 Apr 6. RNA Biol. 2009. PMID: 19246988 Review. - [Misregulation of P-TEFb activity: pathological consequences].
Muniz L, Kiss T, Egloff S. Muniz L, et al. Med Sci (Paris). 2012 Feb;28(2):200-5. doi: 10.1051/medsci/2012282019. Epub 2012 Feb 27. Med Sci (Paris). 2012. PMID: 22377309 Review. French.
Cited by
- Stressing out over long noncoding RNA.
Audas TE, Lee S. Audas TE, et al. Biochim Biophys Acta. 2016 Jan;1859(1):184-91. doi: 10.1016/j.bbagrm.2015.06.010. Epub 2015 Jul 2. Biochim Biophys Acta. 2016. PMID: 26142536 Free PMC article. Review. - An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb.
Kobbi L, Demey-Thomas E, Braye F, Proux F, Kolesnikova O, Vinh J, Poterszman A, Bensaude O. Kobbi L, et al. Proc Natl Acad Sci U S A. 2016 Nov 8;113(45):12721-12726. doi: 10.1073/pnas.1612331113. Epub 2016 Oct 25. Proc Natl Acad Sci U S A. 2016. PMID: 27791144 Free PMC article. - Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity?
Bensaude O. Bensaude O. Transcription. 2011 May;2(3):103-108. doi: 10.4161/trns.2.3.16172. Transcription. 2011. PMID: 21922053 Free PMC article. - Role of noncoding RNAs in the regulation of P-TEFb availability and enzymatic activity.
Napolitano G, Lania L, Majello B. Napolitano G, et al. Biomed Res Int. 2014;2014:643805. doi: 10.1155/2014/643805. Epub 2014 Feb 19. Biomed Res Int. 2014. PMID: 24701579 Free PMC article. Review. - A Cyclin T1 point mutation that abolishes positive transcription elongation factor (P-TEFb) binding to Hexim1 and HIV tat.
Verstraete N, Kuzmina A, Diribarne G, Nguyen VT, Kobbi L, Ludanyi M, Taube R, Bensaude O. Verstraete N, et al. Retrovirology. 2014 Jul 1;11:50. doi: 10.1186/1742-4690-11-50. Retrovirology. 2014. PMID: 24985203 Free PMC article.
References
- Bannai, H., K. Fukatsu, A. Mizutani, T. Natsume, S. Iemura, T. Ikegami, T. Inoue, and K. Mikoshiba. 2004. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279:53427-53434. - PubMed
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
- Bonnal, S., F. Pileur, C. Orsini, F. Parker, F. Pujol, A. C. Prats, and S. Vagner. 2005. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 280:4144-4153. - PubMed
- Byers, S. A., J. P. Price, J. J. Cooper, Q. Li, and D. H. Price. 2005. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 280:16360-16367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials