Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins - PubMed (original) (raw)
Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins
Graydon B Gonsalvez et al. J Cell Biol. 2007.
Abstract
Small nuclear ribonucleoproteins (snRNPs) are core components of the spliceosome. The U1, U2, U4, and U5 snRNPs each contain a common set of seven Sm proteins. Three of these Sm proteins are posttranslationally modified to contain symmetric dimethylarginine (sDMA) residues within their C-terminal tails. However, the precise function of this modification in the snRNP biogenesis pathway is unclear. Several lines of evidence suggest that the methyltransferase protein arginine methyltransferase 5 (PRMT5) is responsible for sDMA modification of Sm proteins. We found that in human cells, PRMT5 and a newly discovered type II methyltransferase, PRMT7, are each required for Sm protein sDMA modification. Furthermore, we show that the two enzymes function nonredundantly in Sm protein methylation. Lastly, we provide in vivo evidence demonstrating that Sm protein sDMA modification is required for snRNP biogenesis in human cells.
Figures
Figure 1.
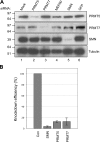
siRNA treatment of PRMT5, PRMT7, MEP50, and SMN. (A) HeLa cells were transfected with siRNAs targeting PRMT5 (lane 2), PRMT7 (lane 3), MEP50 (lane 4), and SMN (lane 5). As a control, cells were untransfected (mock; lane 1) or transfected with siRNAs against GFP (lane 6). 72 h after transfection, lysates were prepared and probed with the indicated antibodies. (B) The protein levels from three separate experiments were quantified. The protein levels were normalized to tubulin and graphed as a fraction of the mock transfection. Error bars represent SD.
Figure 2.
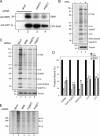
PRMT5 and PRMT7 are required for sDMA modification of Sm proteins. (A) Lysates from mock-treated (lane 1) and PRMT5 (lane 2), PRMT7 (lane 3), MEP50 (lane 4), and SMN (lane 5) siRNA–treated cells were probed with anti-sDMA antibodies SYM10 (top), Y12 (middle), and SYM11 (bottom right). The lysates were probed with tubulin to verify load (bottom left). The open circles represent proteins that require PRMT5, MEP50, and PRMT7 for their methylation. The closed circles represent proteins that require PRMT5 and MEP50 but not PRMT7 for their methylation. The star represents a protein whose methylation status appears unaffected upon the depletion of PRMT5, MEP50, or PRMT7. (B) HeLa cells were treated with siRNAs against PRMT5 (lane 2), PRMT7 (lane 3), or both (lane 4). GFP siRNAs were used as a control (lanes 1–3). Lysates were prepared and analyzed by blotting with the indicated antibodies. (C) HeLa cells were depleted of PRMT5 (lanes 2 and 3). GFP siRNAs were used as a control (lane 1). A FLAG-PRMT7 overexpression construct was transfected into PRMT5-depleted cells (lane 3). Empty vector was transfected as a control (lanes 1 and 2). Lysates were prepared and analyzed by blotting with the indicated antibodies. The asterisk represents a protein that cross reacts with the PRMT7 antibody. (D) 1 μg GST (lanes 1 and 2) or GST-SmD3 C-terminal tail (GST-D3tail; lanes 3 and 4) immobilized on glutathione–Sepharose beads was incubated for 1 h with total HeLa lysates from untreated cells (lanes 1 and 3) or cells treated with MTA (750 μM for 24 h; lanes 2 and 4). The bound proteins were eluted by boiling in SDS buffer and analyzed by blotting using the indicated antibodies.
Figure 3.
sDMA modification of Sm proteins is required for efficient snRNP biogenesis. (A) HeLa cells stably expressing CFP-SmB were depleted of PRMT5 (lane 3) or PRMT7 (lane 4) using RNAi. Mock-treated cells were used as a control (lanes 1 and 2). 72 h after transfection, the cells were harvested, cytoplasmic fractions were prepared, and CFP-SmB was immunoprecipitated using polyclonal GFP antibodies (lanes 2–4). To verify binding specificity, a control myc antibody was used (lane 1). The level of coprecipitating SMN was examined (top). The immunoprecipitation efficiency was verified using a monoclonal GFP antibody (bottom). (B) Cells treated for 24 h with either 750 μM MTA (lane 3) or DMSO (lanes 1 and 2) were pulse labeled with [35S]methionine and [35S]cysteine for 1 h. Subsequently, complete media was added to the cells for a 30-min chase. The cells were harvested, and snRNPs were immunoprecipitated using anti-TMG antibody–coated beads (lanes 2 and 3). Protein A beads were used to demonstrate binding specificity (lane 1). Representative samples were also probed with the tubulin antibody (bottom) to serve as a loading control. The abundant copurifying U1 snRNP proteins are indicated. (C) Cells depleted of SMN (lane 3), PRMT5 (lane 4), or PRMT7 (lane 5) were used in the pulse-chase assay described in A using similar controls. (D) The precipitated levels of the abundant snRNP proteins from three separate RNAi/snRNP assembly experiments were quantified and graphed as a fraction of the mock transfection. Error bars represent SD. (E) HeLa cells stably expressing CFP-SmB were depleted of SMN (lane 3), PRMT5 (lane 4), or PRMT7 (lane 5). Mock-treated cells were used as a control (lanes 1 and 2). The cells were pulsed with [32P]orthophosphate for 2 h. The cells were harvested, total lysates were prepared, and CFP-SmB was immunoprecipitated. Protein A beads were used to demonstrate binding specificity (lane 1). The associated snRNAs were extracted and examined by autoradiography.
Figure 4.
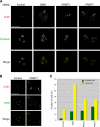
Coilin is found in nucleolar caps in cells depleted of SMN, PRMT5, or PRMT7. (A) Control cells as well as cells depleted of SMN, PRMT5, or PRMT7 were processed for immunofluoresence using antibodies against coilin (red) and fibrillarin (green). The arrows indicate the nucleolar capping phenotype. The images in A and B represent the maximum projection of a z stack that has been deconvolved. (B) Cells depleted of PRMT5 and PRMT7 were processed for immunofluoresence using antibodies against coilin (red) and SMN (green). PRMT7-depleted cells are shown. Note that the coilin nucleolar caps (arrows) also contain SMN. (C) Cells depleted of SMN, PRMT5, or PRMT7 were scored for the presence of CBs and also for the coilin nucleolar capping phenotype. The results were graphed as a percentage of the mock-treated cells. A total of 200 cells were scored for each treatment condition. Bars, 10 μM.
Figure 5.
PRMT5, PRMT7, and SMN depletion affects cytoplasmic snRNP assembly. (A) Cells treated for 24 h with either 750 μM MTA (lanes 2 and 4) or DMSO (lanes 1 and 3) were pulse labeled with 35S as described in Fig. 3 A. The cells were harvested, and nuclear and cytoplasmic fractions were prepared. The snRNPs from each fraction were immunoprecipitated using anti-TMG antibody–coated beads. (B) Cells depleted of SMN (lanes 2 and 6), PRMT5 (lanes 3 and 7), or PRMT7 (lanes 4 and 8) were used in the pulse-chase assay. Mock-treated cells were used as a control (lanes 1 and 5). Binding conditions are the same as in A.
Similar articles
- The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance.
Gonsalvez GB, Rajendra TK, Tian L, Matera AG. Gonsalvez GB, et al. Curr Biol. 2006 Jun 6;16(11):1077-89. doi: 10.1016/j.cub.2006.04.037. Curr Biol. 2006. PMID: 16753561 - Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster.
Gonsalvez GB, Praveen K, Hicks AJ, Tian L, Matera AG. Gonsalvez GB, et al. RNA. 2008 May;14(5):878-87. doi: 10.1261/rna.940708. Epub 2008 Mar 27. RNA. 2008. PMID: 18369183 Free PMC article. - Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein.
Brahms H, Meheus L, de Brabandere V, Fischer U, Lührmann R. Brahms H, et al. RNA. 2001 Nov;7(11):1531-42. doi: 10.1017/s135583820101442x. RNA. 2001. PMID: 11720283 Free PMC article. - The SMN complex: an assembly machine for RNPs.
Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. Battle DJ, et al. Cold Spring Harb Symp Quant Biol. 2006;71:313-20. doi: 10.1101/sqb.2006.71.001. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381311 Review. - Deciphering the assembly pathway of Sm-class U snRNPs.
Neuenkirchen N, Chari A, Fischer U. Neuenkirchen N, et al. FEBS Lett. 2008 Jun 18;582(14):1997-2003. doi: 10.1016/j.febslet.2008.03.009. Epub 2008 Mar 17. FEBS Lett. 2008. PMID: 18348870 Review.
Cited by
- A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ, Jin L, Jacques SL, West KA, Lingaraj T, Stickland K, Ribich SA, Raimondi A, Scott MP, Waters NJ, Pollock RM, Smith JJ, Barbash O, Pappalardi M, Ho TF, Nurse K, Oza KP, Gallagher KT, Kruger R, Moyer MP, Copeland RA, Chesworth R, Duncan KW. Chan-Penebre E, et al. Nat Chem Biol. 2015 Jun;11(6):432-7. doi: 10.1038/nchembio.1810. Epub 2015 Apr 27. Nat Chem Biol. 2015. PMID: 25915199 - Altered gene expression, mitochondrial damage and oxidative stress: converging routes in motor neuron degeneration.
Rossi L, Valle C, Carrì MT. Rossi L, et al. Int J Cell Biol. 2012;2012:908724. doi: 10.1155/2012/908724. Epub 2012 May 17. Int J Cell Biol. 2012. PMID: 22675362 Free PMC article. - Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review).
Zhang J, Jing L, Li M, He L, Guo Z. Zhang J, et al. Mol Med Rep. 2019 May;19(5):3963-3971. doi: 10.3892/mmr.2019.10111. Epub 2019 Apr 1. Mol Med Rep. 2019. PMID: 30942418 Free PMC article. Review. - TDRD6 mediates early steps of spliceosome maturation in primary spermatocytes.
Akpınar M, Lesche M, Fanourgakis G, Fu J, Anastassiadis K, Dahl A, Jessberger R. Akpınar M, et al. PLoS Genet. 2017 Mar 6;13(3):e1006660. doi: 10.1371/journal.pgen.1006660. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28263986 Free PMC article. - Biology of the mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities.
Blijlevens M, Li J, van Beusechem VW. Blijlevens M, et al. Int J Mol Sci. 2021 May 12;22(10):5110. doi: 10.3390/ijms22105110. Int J Mol Sci. 2021. PMID: 34065983 Free PMC article. Review.
References
- Bedford, M.T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell. 18:263–272. - PubMed
- Boisvert, F.M., J. Cote, M.C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics. 2:1319–1330. - PubMed
- Brahms, H., J. Raymackers, A. Union, F. de Keyser, L. Meheus, and R. Lührmann. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275:17122–17129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32-HD007104/HD/NICHD NIH HHS/United States
- R01 GM053034/GM/NIGMS NIH HHS/United States
- S10 RR021228/RR/NCRR NIH HHS/United States
- S10-RR021228/RR/NCRR NIH HHS/United States
- R01 NS041617/NS/NINDS NIH HHS/United States
- R01-GM53034/GM/NIGMS NIH HHS/United States
- S10-RR017980/RR/NCRR NIH HHS/United States
- S10 RR017980/RR/NCRR NIH HHS/United States
- R01-NS41617/NS/NINDS NIH HHS/United States
- Wellcome Trust/United Kingdom
- T32 HD007104/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases