Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation - PubMed (original) (raw)
. 2007 Sep 13;449(7159):248-51.
doi: 10.1038/nature06146. Epub 2007 Aug 22.
Affiliations
- PMID: 17713477
- PMCID: PMC2712830
- DOI: 10.1038/nature06146
Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation
Da Jia et al. Nature. 2007.
Abstract
Genetic imprinting, found in flowering plants and placental mammals, uses DNA methylation to yield gene expression that is dependent on the parent of origin. DNA methyltransferase 3a (Dnmt3a) and its regulatory factor, DNA methyltransferase 3-like protein (Dnmt3L), are both required for the de novo DNA methylation of imprinted genes in mammalian germ cells. Dnmt3L interacts specifically with unmethylated lysine 4 of histone H3 through its amino-terminal PHD (plant homeodomain)-like domain. Here we show, with the use of crystallography, that the carboxy-terminal domain of human Dnmt3L interacts with the catalytic domain of Dnmt3a, demonstrating that Dnmt3L has dual functions of binding the unmethylated histone tail and activating DNA methyltransferase. The complexed C-terminal domains of Dnmt3a and Dnmt3L showed further dimerization through Dnmt3a-Dnmt3a interaction, forming a tetrameric complex with two active sites. Substitution of key non-catalytic residues at the Dnmt3a-Dnmt3L interface or the Dnmt3a-Dnmt3a interface eliminated enzymatic activity. Molecular modelling of a DNA-Dnmt3a dimer indicated that the two active sites are separated by about one DNA helical turn. The C-terminal domain of Dnmt3a oligomerizes on DNA to form a nucleoprotein filament. A periodicity in the activity of Dnmt3a on long DNA revealed a correlation of methylated CpG sites at distances of eight to ten base pairs, indicating that oligomerization leads Dnmt3a to methylate DNA in a periodic pattern. A similar periodicity is observed for the frequency of CpG sites in the differentially methylated regions of 12 maternally imprinted mouse genes. These results suggest a basis for the recognition and methylation of differentially methylated regions in imprinted genes, involving the detection of both nucleosome modification and CpG spacing.
Figures
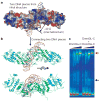
Figure 1. Structure of the Dnmt3a–Dnmt3L (3a–3L) C-terminal domain complex
a, Side view of the tetramer Dnmt3L (blue)–Dnmt3a (green) –Dnmt3a (green)–Dnmt3L (blue), with the bound AdoHcy shown as a stick model. The inset shows the two pairs of phenylalanine residues (F728 and F768 of Dnmt3a, and F261 and F301 of Dnmt3L) in the centre of the Dnmt3a–Dnmt3L interface. b, Top view showing the Dnmt3a–Dnmt3a interface with two pairs of salt bridges formed between R881 and D872 (enlarged). c, Activities of Dnmt3a2 and its point mutant F728A in the presence and absence of Dnmt3L. Error bars represent s.d. calculated from two independent experiments. d, A network of polar interactions between two Dnmt3a molecules (coloured blue and purple) involves R881–D872, D872–H869, H869–E851 and E851–R881.
Figure 2. Model of Dnmt3a–Dnmt3L tetramer with DNA
a, Surface representation of the Dnmt3a–Dnmt3L tetramer, with two short DNA molecules adopted by superimposition of the _Hha_I–DNA complex structure onto individual Dnmt3a-C. b, Two views of the Dnmt3a–Dnmt3L tetramer with one contiguous curved DNA molecule, approximately 25 base pairs in length, covering two active sites. c, Cooperative formation of large protein–DNA complexes (red arrow) after incubation of a 146-base-pair DNA with increasing concentrations of Dnmt3a-C or Dnmt3a-C–Dnmt3L-C complex. The lower band (black arrow) corresponds to one Dnmt3L-C bound to the DNA as a result of the presence of some free Dnmt3L-C in the mixture (Supplementary Fig. 5a).
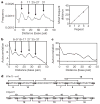
Figure 3. Periodicity of Dnmt3a-C activity on long DNA substrates
a, Two DNA fragments (from λ phage (top; 520 base pairs, 40 CpGs) and the CpG island upstream of the human SUHW1 gene (bottom; 420 base pairs, 56 CpGs)) were used for methylation. A total of 119 clones (64 from λ phage, 55 from the human CpG island) were sequenced to determine their methylation pattern. b, Autocorrelation of CpG methylation. By averaging two substrates, the P values for the correlation factors observed for distances of eight to ten base pairs (arrow) are in the range (3–7) × 10−4. This periodicity is detectable for correlation of methylation of individual CpG sites as well as for correlation of the absence of methylation at two sites (Supplementary Fig. 6).
Figure 4. Periodicity of CpG sites in maternally imprinted DMRs
a, Frequencies of CpG pairs at a given distance with respect to all pairs, for 11 maternally imprinted DMRs. The slope of the graph in the inset is 9.5. b, The autocorrelation of peaks for 11 maternally imprinted DMRs. One sequence (Impact) was excluded from the plot because it contains repeats (see d), with each CpG site separated from the second subsequent one by ten base pairs; thus, it follows the same pattern seen by the other 11 DMRs, bringing the total number to 12 DMRs. c, Autocorrelation of peaks for ten CpG islands randomly selected from human chromosome 21. d, A region of DMR sequences of the _U2af1_-rs1 and Impact genes.
Comment in
- Epigenetics: perceptive enzymes.
Ferguson-Smith AC, Greally JM. Ferguson-Smith AC, et al. Nature. 2007 Sep 13;449(7159):148-9. doi: 10.1038/449148a. Nature. 2007. PMID: 17851501 No abstract available.
Similar articles
- DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. Ooi SK, et al. Nature. 2007 Aug 9;448(7154):714-7. doi: 10.1038/nature05987. Nature. 2007. PMID: 17687327 Free PMC article. - Structural insight into autoinhibition and histone H3-induced activation of DNMT3A.
Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Guo X, et al. Nature. 2015 Jan 29;517(7536):640-4. doi: 10.1038/nature13899. Epub 2014 Nov 10. Nature. 2015. PMID: 25383530 - The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a.
Chedin F, Lieber MR, Hsieh CL. Chedin F, et al. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16916-21. doi: 10.1073/pnas.262443999. Epub 2002 Dec 12. Proc Natl Acad Sci U S A. 2002. PMID: 12481029 Free PMC article. - Multimerization of the dnmt3a DNA methyltransferase and its functional implications.
Jeltsch A, Jurkowska RZ. Jeltsch A, et al. Prog Mol Biol Transl Sci. 2013;117:445-64. doi: 10.1016/B978-0-12-386931-9.00016-7. Prog Mol Biol Transl Sci. 2013. PMID: 23663978 Review. - Molecular coupling of DNA methylation and histone methylation.
Hashimoto H, Vertino PM, Cheng X. Hashimoto H, et al. Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review.
Cited by
- Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - Clonal hematopoiesis and hematological malignancy.
Dunn WG, McLoughlin MA, Vassiliou GS. Dunn WG, et al. J Clin Invest. 2024 Oct 1;134(19):e180065. doi: 10.1172/JCI180065. J Clin Invest. 2024. PMID: 39352393 Free PMC article. Review. - Closing in on human methylation-the versatile family of seven-β-strand (METTL) methyltransferases.
Falnes PØ. Falnes PØ. Nucleic Acids Res. 2024 Oct 28;52(19):11423-11441. doi: 10.1093/nar/gkae816. Nucleic Acids Res. 2024. PMID: 39351878 Free PMC article. Review. - The Role of DNMT Methyltransferases and TET Dioxygenases in the Maintenance of the DNA Methylation Level.
Davletgildeeva AT, Kuznetsov NA. Davletgildeeva AT, et al. Biomolecules. 2024 Sep 4;14(9):1117. doi: 10.3390/biom14091117. Biomolecules. 2024. PMID: 39334883 Free PMC article. Review. - DNA-binding proteins from MBD through ZF to BEN: recognition of cytosine methylation status by one arginine with two conformations.
Zhang X, Blumenthal RM, Cheng X. Zhang X, et al. Nucleic Acids Res. 2024 Oct 28;52(19):11442-11454. doi: 10.1093/nar/gkae832. Nucleic Acids Res. 2024. PMID: 39329271 Free PMC article.
References
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. - PubMed
- Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. - PubMed
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases