Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent - PubMed (original) (raw)
Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent
Jennifer Jemc et al. Dev Biol. 2007.
Abstract
Drosophila eye specification and development relies on a collection of transcription factors termed the retinal determination gene network (RDGN). Two members of this network, Eyes absent (EYA) and Sine oculis (SO), form a transcriptional complex in which EYA provides the transactivation function while SO provides the DNA binding activity. EYA also functions as a protein tyrosine phosphatase, raising the question of whether transcriptional output is dependent or independent of phosphatase activity. To explore this, we used microarrays together with binding site analysis, quantitative real-time PCR, chromatin immunoprecipitation, genetics and in vivo expression analysis to identify new EYA-SO targets. In parallel, we examined the expression profiles of tissue expressing phosphatase mutant eya and found that reducing phosphatase activity did not globally impair transcriptional output. Among the targets identified by our analysis was the cell cycle regulatory gene, string (stg), suggesting that EYA and SO may influence cell proliferation through transcriptional regulation of stg. Future investigation into the regulation of stg and other EYA-SO targets identified in this study will help elucidate the transcriptional circuitries whereby output from the RDGN integrates with other signaling inputs to coordinate retinal development.
Figures
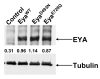
Figure 1
Eya transgenes are expressed at equivalent levels. Western blot of head lysates from flies overexpressing _eya_WT, _eya_D493N, and _eya_E728Q.
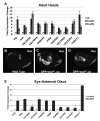
Figure 2
Overexpression of eya induces expression of putative target genes. (A) Quantitative RT-PCR analysis of target expression in adult heads overexpressing eyaWT and eyaMUT under control of the HSP-Gal4 driver compared to adult heads of flies carrying the driver alone. (B-D) Dac antibody staining in wing imaginal discs. (B) Wild type. (C) DPP-Gal4 driving coexpression of eyaWT and so transgenes. (D) Wing imaginal disc overexpressing eyaMUT and so under the control of the DPP-Gal4 driving coexpression of the phosphatase dead eyaEQ and so transgenes. (C-D) Dac expression is induced strongly by eyaWT and eyaMUT when overexpressed with so. (E) Quantitative RT-PCR of target expression in third instar larval eye-antennal imaginal discs overexpressing eyaWT under the control of the GMR-Gal4 driver compared to discs only carrying the driver alone.
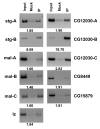
Figure 3
Chromatin immunoprecipitation analysis of SO association with genomic regions containing predicted SO binding sites. PCR amplified product obtained from input chromatin is in the first lane, from mock treated sample incubated with pre-immune serum is in the second lane, and from sample immunoprecipitated using the SO antibody is in the third lane. Chromatin enrichment in the IP sample over the mock treated sample is given below each panel.
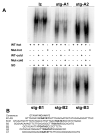
Figure 4
SO binding analysis. (A) Electrophoretic mobility shift assays with recombinant GST-full-length SO on double-stranded oligonucleotides containing predicted wild type or mutant SO binding sites. (B) Alignment of predicted SO binding sites highlights the core consensus sequence.
Figure 5
Heterozyosity for stg reduces the frequency of ectopic eye induction upon eya misexpression under the DPP-Gal4 driver.
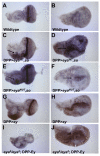
Figure 6
EYA and SO regulation of stg expression. (A-H) In situ hybridization for stg. Wild type eyeantennal (A) or wing imaginal disc (B) show strong stg. Eye-antennal (C) or wing (D) imaginal discs overexpressing eyaWT and so under the control of the DPP-Gal4 driver. In both tissues stg expression is induced in the region where eya and so are being overexpressed. Eye-antennal (E) and wing (F) imaginal discs in which eyaMUT and so are overexpressed under the control of the DPP-Gal4 driver demonstrate ectopic stg expression. Arrows in C-F indicate regions of increased stg expression. Overexpression of ey using a DPP-Gal4 driver induces stg expression in eye-antennal (G) and wing imaginal discs (H). Discs overexpressing ey, but homozygous for the eya2 allele no longer exhibit induction of stg expression (I, J). In eya2/eya2; DPP-Gal4/ UAS-ey, wing discs significant folding is observed, which results in the darker stg staining in the region of ey overexpression but does not reflect an overall increase in stg expression.
Similar articles
- Distinct Biochemical Activities of Eyes absent During Drosophila Eye Development.
Jin M, Mardon G. Jin M, et al. Sci Rep. 2016 Mar 16;6:23228. doi: 10.1038/srep23228. Sci Rep. 2016. PMID: 26980695 Free PMC article. - Mutations that impair Eyes absent tyrosine phosphatase activity in vitro reduce robustness of retinal determination gene network output in Drosophila.
Davis TL, Hoi CSL, Rebay I. Davis TL, et al. PLoS One. 2017 Nov 6;12(11):e0187546. doi: 10.1371/journal.pone.0187546. eCollection 2017. PLoS One. 2017. PMID: 29108015 Free PMC article. - Antagonistic regulation of the second mitotic wave by Eyes absent-Sine oculis and Combgap coordinates proliferation and specification in the Drosophila retina.
Davis TL, Rebay I. Davis TL, et al. Development. 2017 Jul 15;144(14):2640-2651. doi: 10.1242/dev.147231. Epub 2017 Jun 15. Development. 2017. PMID: 28619818 Free PMC article. - Regulation of cancer stem cell properties by SIX1, a member of the PAX-SIX-EYA-DACH network.
Kingsbury TJ, Kim M, Civin CI. Kingsbury TJ, et al. Adv Cancer Res. 2019;141:1-42. doi: 10.1016/bs.acr.2018.12.001. Epub 2019 Jan 16. Adv Cancer Res. 2019. PMID: 30691681 Review. - The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription.
Jemc J, Rebay I. Jemc J, et al. Annu Rev Biochem. 2007;76:513-38. doi: 10.1146/annurev.biochem.76.052705.164916. Annu Rev Biochem. 2007. PMID: 17341163 Review.
Cited by
- Nemo phosphorylates Eyes absent and enhances output from the Eya-Sine oculis transcriptional complex during Drosophila retinal determination.
Morillo SA, Braid LR, Verheyen EM, Rebay I. Morillo SA, et al. Dev Biol. 2012 May 1;365(1):267-76. doi: 10.1016/j.ydbio.2012.02.030. Epub 2012 Feb 25. Dev Biol. 2012. PMID: 22394486 Free PMC article. - The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells.
Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. Pandey RN, et al. Oncogene. 2010 Jun 24;29(25):3715-22. doi: 10.1038/onc.2010.122. Epub 2010 Apr 26. Oncogene. 2010. PMID: 20418914 Free PMC article. - Multilevel regulation of the glass locus during Drosophila eye development.
Fritsch C, Bernardo-Garcia FJ, Humberg TH, Mishra AK, Miellet S, Almeida S, Frochaux MV, Deplancke B, Huber A, Sprecher SG. Fritsch C, et al. PLoS Genet. 2019 Jul 12;15(7):e1008269. doi: 10.1371/journal.pgen.1008269. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31299050 Free PMC article. - Context dependence of proneural bHLH proteins.
Powell LM, Jarman AP. Powell LM, et al. Curr Opin Genet Dev. 2008 Oct;18(5):411-7. doi: 10.1016/j.gde.2008.07.012. Epub 2008 Sep 7. Curr Opin Genet Dev. 2008. PMID: 18722526 Free PMC article. Review. - Eye selector logic for a coordinated cell cycle exit.
Lopes CS, Casares F. Lopes CS, et al. PLoS Genet. 2015 Feb 19;11(2):e1004981. doi: 10.1371/journal.pgen.1004981. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25695251 Free PMC article.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–64. - PubMed
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42.
- Aligianis IA, Johnson CA, Gissen P, Chen D, Hampshire D, Hoffmann K, Maina EN, Morgan NV, Tee L, Morton J, Ainsworth JR, Horn D, Rosser E, Cole TR, Stolte-Dijkstra I, Fieggen K, Clayton-Smith J, Megarbane A, Shield JP, Newbury-Ecob R, Dobyns WB, Graham JM, Jr., Kjaer KW, Warburg M, Bond J, Trembath RC, Harris LW, Takai Y, Mundlos S, Tannahill D, Woods CG, Maher ER. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet. 2005;37:221–3. - PubMed
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–88. - PubMed
- Amrani L, Primus J, Glatigny A, Arcangeli L, Scazzocchio C, Finnerty V. Comparison of the sequences of the Aspergillus nidulans hxB and Drosophila melanogaster ma-l genes with nifS from Azotobacter vinelandii suggests a mechanism for the insertion of the terminal sulphur atom in the molybdopterin cofactor. Mol Microbiol. 2000;38:114–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY012549/EY/NEI NIH HHS/United States
- R01 EY012549-03/EY/NEI NIH HHS/United States
- R01 EY12549/EY/NEI NIH HHS/United States
- R01 EY012549-06/EY/NEI NIH HHS/United States
- R01 EY012549-04/EY/NEI NIH HHS/United States
- R01 EY012549-05/EY/NEI NIH HHS/United States
- R01 EY012549-07/EY/NEI NIH HHS/United States
- R01 EY012549-05S1/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases