Patterns of damage in genomic DNA sequences from a Neandertal - PubMed (original) (raw)
Patterns of damage in genomic DNA sequences from a Neandertal
Adrian W Briggs et al. Proc Natl Acad Sci U S A. 2007.
Abstract
High-throughput direct sequencing techniques have recently opened the possibility to sequence genomes from Pleistocene organisms. Here we analyze DNA sequences determined from a Neandertal, a mammoth, and a cave bear. We show that purines are overrepresented at positions adjacent to the breaks in the ancient DNA, suggesting that depurination has contributed to its degradation. We furthermore show that substitutions resulting from miscoding cytosine residues are vastly overrepresented in the DNA sequences and drastically clustered in the ends of the molecules, whereas other substitutions are rare. We present a model where the observed substitution patterns are used to estimate the rate of deamination of cytosine residues in single- and double-stranded portions of the DNA, the length of single-stranded ends, and the frequency of nicks. The results suggest that reliable genome sequences can be obtained from Pleistocene organisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
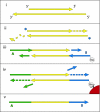
Fig. 1.
The 454 library preparation process. Double-stranded DNA molecules (i) (yellow) are made blunt-ended by T4 DNA polymerase, 5′-phosphorylated (stars) by T4 polynucleotide kinase (ii) and ligated to one strand of nonphosphorylated double-stranded adaptors A (green) and B (blue) (iii). Ligation products carrying the biotinylated B adaptor are captured on Streptavidin beads (red), and the strand-displacing Bst DNA polymerase is used to extend the nicks between adaptors and template (iv). The DNA strands are then denatured, releasing the A-to-B strands (v), which are isolated and used as templates for emulsion PCR.
Fig. 2.
Base composition at ends of Neandertal DNA sequences. The base composition of the human reference sequence is plotted as a function of distance from 5′- and 3′-ends of Neandertal sequences.
Fig. 3.
Misincorporation patterns in Neandertal DNA sequences. The frequencies of the 12 possible mismatches are plotted as a function of distance from 5′- and 3′-ends. At each position, the substitution frequency, e.g., C-T, is calculated as the proportion of human reference sequence positions carrying C where the 454 sequence is T. The 10 5′- and 10 3′-most nucleotides were removed from the 3′- and 5′-graphs, respectively.
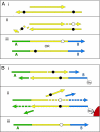
Fig. 4.
Miscoding lesions and the 454 process. During preparation of templates for 454 sequencing, the ends of DNA fragments are first repaired by T4 DNA polymerase (A), and in a later step linkers are filled in by Bst DNA polymerase (B). During blunt-end repair by T4 DNA polymerase (A), miscoding lesions (black circles) on 3′-overhanging ends are removed, whereas miscoding lesions on 5′-overhangs result in complementary misincorporations (white circles) in the resultant 454 sequences. Similarly, extension by the strand-displacing Bst DNA polymerase (B) causes miscoding lesions in the template DNA downstream of nicks or gaps to result in complementary misincorporations in the sequences generated.
Similar articles
- Ancient DNA damage.
Dabney J, Meyer M, Pääbo S. Dabney J, et al. Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a012567. doi: 10.1101/cshperspect.a012567. Cold Spring Harb Perspect Biol. 2013. PMID: 23729639 Free PMC article. Review. - DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA.
Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. Hofreiter M, et al. Nucleic Acids Res. 2001 Dec 1;29(23):4793-9. doi: 10.1093/nar/29.23.4793. Nucleic Acids Res. 2001. PMID: 11726688 Free PMC article. - Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA.
Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, Antipina EE, Baryshnikov GF, Kuzmin YV, Vasilevski AA, Wuenschell GE, Termini J, Hofreiter M, Jaenicke-Després V, Pääbo S. Stiller M, et al. Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13578-84. doi: 10.1073/pnas.0605327103. Epub 2006 Aug 25. Proc Natl Acad Sci U S A. 2006. PMID: 16938852 Free PMC article. - Genomic sequencing of Pleistocene cave bears.
Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, Rabeder G, Krause J, Detter JC, Pääbo S, Rubin EM. Noonan JP, et al. Science. 2005 Jul 22;309(5734):597-9. doi: 10.1126/science.1113485. Epub 2005 Jun 2. Science. 2005. PMID: 15933159 - Ancient genomes.
Hoelzel AR. Hoelzel AR. Genome Biol. 2005;6(12):239. doi: 10.1186/gb-2005-6-12-239. Epub 2005 Dec 1. Genome Biol. 2005. PMID: 16356273 Free PMC article. Review.
Cited by
- Ancient DNA damage.
Dabney J, Meyer M, Pääbo S. Dabney J, et al. Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a012567. doi: 10.1101/cshperspect.a012567. Cold Spring Harb Perspect Biol. 2013. PMID: 23729639 Free PMC article. Review. - Estimating molecular preservation of the intestinal microbiome via metagenomic analyses of latrine sediments from two medieval cities.
Sabin S, Yeh HY, Pluskowski A, Clamer C, Mitchell PD, Bos KI. Sabin S, et al. Philos Trans R Soc Lond B Biol Sci. 2020 Nov 23;375(1812):20190576. doi: 10.1098/rstb.2019.0576. Epub 2020 Oct 5. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 33012229 Free PMC article. - Unravelling reference bias in ancient DNA datasets.
Dolenz S, van der Valk T, Jin C, Oppenheimer J, Sharif MB, Orlando L, Shapiro B, Dalén L, Heintzman PD. Dolenz S, et al. Bioinformatics. 2024 Jul 1;40(7):btae436. doi: 10.1093/bioinformatics/btae436. Bioinformatics. 2024. PMID: 38960861 Free PMC article. - Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA.
Gansauge MT, Meyer M. Gansauge MT, et al. Nat Protoc. 2013 Apr;8(4):737-48. doi: 10.1038/nprot.2013.038. Epub 2013 Mar 14. Nat Protoc. 2013. PMID: 23493070 - The genetics of an early Neolithic pastoralist from the Zagros, Iran.
Gallego-Llorente M, Connell S, Jones ER, Merrett DC, Jeon Y, Eriksson A, Siska V, Gamba C, Meiklejohn C, Beyer R, Jeon S, Cho YS, Hofreiter M, Bhak J, Manica A, Pinhasi R. Gallego-Llorente M, et al. Sci Rep. 2016 Aug 9;6:31326. doi: 10.1038/srep31326. Sci Rep. 2016. PMID: 27502179 Free PMC article.
References
- Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Nat Rev Genet. 2001;2:353–359. - PubMed
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Annu Rev Genet. 2004;38:645–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources