Design of extended short hairpin RNAs for HIV-1 inhibition - PubMed (original) (raw)
Design of extended short hairpin RNAs for HIV-1 inhibition
Ying Poi Liu et al. Nucleic Acids Res. 2007.
Abstract
RNA interference (RNAi) targeted towards viral mRNAs is widely used to block virus replication in mammalian cells. The specific antiviral RNAi response can be induced via transfection of synthetic small interfering RNAs (siRNAs) or via intracellular expression of short hairpin RNAs (shRNAs). For HIV-1, both approaches resulted in profound inhibition of virus replication. However, the therapeutic use of a single siRNA/shRNA appears limited due to the rapid emergence of RNAi-resistant escape viruses. These variants contain deletions or point mutations within the target sequence that abolish the antiviral effect. To avoid escape from RNAi, the virus should be simultaneously targeted with multiple shRNAs. Alternatively, long hairpin RNAs can be used from which multiple effective siRNAs may be produced. In this study, we constructed extended shRNAs (e-shRNAs) that encode two effective siRNAs against conserved HIV-1 sequences. Activity assays and RNA processing analyses indicate that the positioning of the two siRNAs within the hairpin stem is critical for the generation of two functional siRNAs. E-shRNAs that are efficiently processed into two effective siRNAs showed better inhibition of virus production than the poorly processed e-shRNAs, without inducing the interferon response. These results provide building principles for the design of multi-siRNA hairpin constructs.
Figures
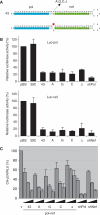
Figure 1.
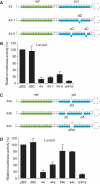
Silencing activities of extended hairpin RNAs depend on the position of the active siRNA in the stem. (A) Schematic of the 39 bp extended hairpin RNAs and the 21 bp original shGag. Extended hairpins against gag were constructed with the effective gag siRNA (antisense strand boxed) at the base (39B), center (39C) and top (39T). (B) Luciferase reporters containing a broad 50 nt or a precise 19 nt gag target. (C) Reporter constructs (100 ng) and the control renilla luciferase expression plasmid (1 ng) were used to co-transfect 293T cells together with different amounts of 39 bp extended hairpin RNA constructs and the original shGag. Firefly and renilla luciferase activity was measured 2 days post-transfection and the ratio was used to calculate the relative luciferase activity. Luciferase activity in the absence of inhibitor was set at 100%. The mean values obtained in three independent experiments are shown.
Figure 2.
Optimization of a bifunctional extended shRNA. (A) Schematic of the e-shRNAs pol-nef and nef-pol of 40 bp and the length variants of 41–44 bp. The antisense strand of the siRNAs against pol and nef are marked in blue and green, respectively. (B) Luciferase reporter constructs containing 50 nt HIV-1 pol (Luc-pol) or nef (Luc-nef) sequences, with the 19 nt target in the center. (C) 293T cells were co-transfected with 100 ng of the Luc-pol (left) or Luc-nef (right) reporter, 1 ng of the renilla luciferase plasmid and 10 ng of the e-shRNAs Relative luciferase activities were determined from the firefly and the control renilla luciferase expression ratios. Luciferase expression in the presence of pBS was set at 100%. The 39C construct against HIV-1 gag was used as a negative control. The original shPol and shNef were included as positive controls. Averages and standard deviations represent three independent transfections.
Figure 3.
Variants of nef-pol 44 to test the specificity of the e-shRNA design. (A) The original nef-pol 44 hairpin and two variants that contain either a 1 or 2 nt deletion (44-1/44-2) in the passenger/sense strand. These variants have a reduced stem length of 1 bp (43 bp) or 2 bp (42 bp). (B) 293T cells were co-transfected with 10 ng of the indicated hairpin constructs, 100 ng of the luciferase reporter and 1 ng of the renilla luciferase plasmid. Two days post-transfection, knockdown efficiencies were determined using the firefly and renilla luciferase expression ratios. (C) Three additional mutant hairpins were made that contain either one (44a) or two (44b and 44c) deletions in the guide/antisense strand of the hairpin. (D) 293T cells were co-transfected with 10 ng of 44a, 44b and 44c, 100 ng of the Luc-pol reporter and 1 ng of the renilla luciferase plasmid. Two days post-transfection, firefly and renilla luciferase expression was measured and the ratio was used to determine the relative luciferase activity. Luciferase expression in the presence of pBS was set at 100%. Averages and standard deviations represent three independent transfections.
Figure 4.
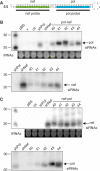
Northern blot analysis of the e-shRNAs derived siRNAs. (A) The nef-pol 44 hairpin with the probes to detect siRNAs against pol and nef. (B) Small RNAs were purified from 293T cells transfected with the pol-nef e-shRNA variants 40–44 for detection of siRNAs pol and nef. pBluescript (pBS), the empty vector (v) and the irrelevant 39C hairpin against gag (ctr) were used as negative controls. (C) Detection of siRNAs nef and pol derived from nef-pol e-shRNAs transfected 293T cells. pBS and the irrelevant 39C hairpin against gag (ctr) were used as negative controls. Ethidium bromide staining of tRNAs serves as sample loading control. Ethidium bromide stainings were shown for the upper blot of Figure 4B and C. Stainings were performed for all northern blots and a similar picture was observed.
Figure 5.
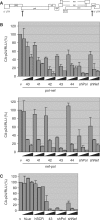
Inhibition of HIV-1 production by e-shRNA constructs. (A) The HIV-1 genome and the position of the target sequences for the e-shRNA derived siRNAs. (B) 293T cells were co-transfected with 250 ng of the HIV-1 pLAI, 1 ng of renilla luciferase plasmid and 2.5, 10 or 25 ng of the e-shRNA constructs. Two days post-transfection, inhibition of HIV-1 production was determined by measuring CA-p24 levels in the culture supernatant. CA-p24 levels were normalized to the renilla luciferase activities. The ratio between the CA-p24 level and the renilla luciferase activity in the presence of 25 ng of the empty vector (v) was set at 100%. (C) 293T cells were co-transfected with two control hairpin constructs. One encodes two siRNAs targeting the luciferase gene (hLuc) and the other encodes the scrambled sequence of the 43 bp e-shRNA (hSCR). The nef-pol 43 bp construct was used as a positive control. The level of HIV-1 inhibition was determined as described in B. Averages and standard deviations represent two independent experiments.
Figure 6.
Towards user-friendly e-shRNA variants. (A) A mutation or deletion was introduced in the passenger/sense strand of the pol-nef 43 hairpin, which creates a mismatch in the central stem region. (B) 293T cells were co-transfected with 10 ng of the modified e-shRNAs, 100 ng of the Luc-pol or Luc-nef and 1 ng of the renilla luciferase plasmid. Two days post-transfection relative luciferase activities were determined using the firefly and renilla luciferase expression ratios. Luciferase expression in the presence of pBS was set at 100%. Averages and standard deviations represent three independent transfections. (C) Inhibition of HIV-1 production by the modified e-shRNAs was determined by co-transfection of 293T cells with 2.5, 10 or 25 ng of the hairpins, 250 ng of the HIV-1 pLAI and 1 ng of the renilla luciferase plasmid. CA-p24 levels were measured from the culture supernatant 2 days post-transfection. The ratio between the CA-p24 and the renilla luciferase activity in the presence of 25 ng of the empty vector (v) was set at 100%. Averages and standard deviations represent two independent experiments.
Figure 7.
E-shRNAs do not induce an interferon response. Total RNA was purified from 293T cells transfected with the pol-nef e-shRNA variants 40–44, the 43 bp A mutant, the original shRNAs and a 43 bp hairpin encoding the scrambled sequence of the 43 bp e-shRNA (hSCR). The IFN-β, OAS, MxA and ISG56 expression levels were determined by RT–PCR. β-actin mRNA expression serves as an internal control. Negative controls include the mock (H2O) transfection and the PCR reaction on RNA extracts derived from dsRNA transfected cells that were not subjected to reverse transcription. In vitro transcribed dsRNA (300 bp nef sequences) act as positive control for IFN-β induction. The 200 bp lane of the SmartLadder (Eurogentec) is shown as a size reference. For the OAS amplicon product, the 400 bp lane of the SmartLadder is shown.
Similar articles
- Stringent testing identifies highly potent and escape-proof anti-HIV short hairpin RNAs.
von Eije KJ, ter Brake O, Berkhout B. von Eije KJ, et al. J Gene Med. 2009 Jun;11(6):459-67. doi: 10.1002/jgm.1329. J Gene Med. 2009. PMID: 19384894 - The inhibitory efficacy of RNA POL III-expressed long hairpin RNAs targeted to untranslated regions of the HIV-1 5' long terminal repeat.
Barichievy S, Saayman S, von Eije KJ, Morris KV, Arbuthnot P, Weinberg MS. Barichievy S, et al. Oligonucleotides. 2007 Winter;17(4):419-31. doi: 10.1089/oli.2007.0095. Oligonucleotides. 2007. PMID: 17896874 - Combinatorial RNAi against HIV-1 using extended short hairpin RNAs.
Liu YP, von Eije KJ, Schopman NC, Westerink JT, ter Brake O, Haasnoot J, Berkhout B. Liu YP, et al. Mol Ther. 2009 Oct;17(10):1712-23. doi: 10.1038/mt.2009.176. Epub 2009 Aug 11. Mol Ther. 2009. PMID: 19672247 Free PMC article. - Lentiviral delivery of RNAi effectors against HIV-1.
Liu YP, Berkhout B. Liu YP, et al. Curr Top Med Chem. 2009;9(12):1130-43. doi: 10.2174/156802609789630866. Curr Top Med Chem. 2009. PMID: 19860713 Review. - RNA-interference-based gene therapy approaches to HIV type-1 treatment: tackling the hurdles from bench to bedside.
von Eije KJ, Berkhout B. von Eije KJ, et al. Antivir Chem Chemother. 2009;19(6):221-33. doi: 10.1177/095632020901900602. Antivir Chem Chemother. 2009. PMID: 19641231 Review.
Cited by
- Cassette deletion in multiple shRNA lentiviral vectors for HIV-1 and its impact on treatment success.
Mcintyre GJ, Yu YH, Tran A, Jaramillo AB, Arndt AJ, Millington ML, Boyd MP, Elliott FA, Shen SW, Murray JM, Applegate TL. Mcintyre GJ, et al. Virol J. 2009 Oct 30;6:184. doi: 10.1186/1743-422X-6-184. Virol J. 2009. PMID: 19878571 Free PMC article. - Possible applications for replicating HIV 1 vectors.
Das AT, Jeeninga RE, Berkhout B. Das AT, et al. HIV Ther. 2010 May 1;4(3):361-369. doi: 10.2217/hiv.10.20. HIV Ther. 2010. PMID: 20582153 Free PMC article. - Multiple shRNA combinations for near-complete coverage of all HIV-1 strains.
McIntyre GJ, Groneman JL, Yu YH, Tran A, Applegate TL. McIntyre GJ, et al. AIDS Res Ther. 2011 Jan 13;8(1):1. doi: 10.1186/1742-6405-8-1. AIDS Res Ther. 2011. PMID: 21226969 Free PMC article. - A direct comparison of strategies for combinatorial RNA interference.
Lambeth LS, Van Hateren NJ, Wilson SA, Nair V. Lambeth LS, et al. BMC Mol Biol. 2010 Oct 11;11:77. doi: 10.1186/1471-2199-11-77. BMC Mol Biol. 2010. PMID: 20937117 Free PMC article. - Bone Marrow Gene Therapy for HIV/AIDS.
Herrera-Carrillo E, Berkhout B. Herrera-Carrillo E, et al. Viruses. 2015 Jul 17;7(7):3910-36. doi: 10.3390/v7072804. Viruses. 2015. PMID: 26193303 Free PMC article. Review.
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. - PubMed
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. - PubMed
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. - PubMed
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical