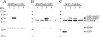Refined characterization of the expression and stability of the SMN gene products - PubMed (original) (raw)
Refined characterization of the expression and stability of the SMN gene products
Jérémie Vitte et al. Am J Pathol. 2007 Oct.
Abstract
Spinal muscular atrophy (SMA) is characterized by degeneration of lower motor neurons and caused by mutations of the SMN1 gene. SMN1 is duplicated in a homologous gene called SMN2, which remains present in patients. SMN has an essential role in RNA metabolism, but its role in SMA pathogenesis remains unknown. Previous studies suggested that in neurons the protein lacking the C terminus (SMN(Delta7)), the major product of the SMN2 gene, had a dominant-negative effect. We generated antibodies specific to SMN(FL) or SMN(Delta7). In transfected cells, the stability of the SMN(Delta7) protein was regulated in a cell-dependent manner. Importantly, whatever the human tissues examined, SMN(Delta7) protein was undetectable because of the instability of the protein, thus excluding a dominant effect of SMN(Delta7) in SMA. A similar decreased level of SMN(FL) was observed in brain and spinal cord samples from human SMA, suggesting that SMN(FL) may have specific targets in motor neurons. Moreover, these data indicate that the vulnerability of motor neurons cannot simply be ascribed to the differential expression or a more dramatic reduction of SMN(FL) in spinal cord when compared with brain tissue. Improving the stability of SMN(Delta7) protein might be envisaged as a new therapeutic strategy in SMA.
Figures
Figure 1
Characterization of new polyclonal antibodies directed against human SMNFL and SMNΔ7 proteins. Immunoblot analysis of proteins extracted from HeLa cells transfected with a plasmid expressing GFP (lane 1), GFP-SMNFL (lane 2), GFP-SMNΔ7 (lane 3), or from nontransfected HeLa cells (lane 4). The membranes were incubated with hSMNex7-5381 (A), hSMNex8-5699 (B) antisera, or with GFP and tubulin antibodies (C). An asterisk indicates a nonspecific band (see comment on Figure 7).
Figure 2
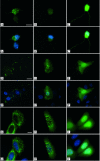
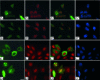
Expression and subcellular localization of SMNFL and SMNΔ7 in various mammalian cell types. Primary cultures of mouse cortical neurons (A–C′), mouse fibroblasts (D–F′), and HeLa cells (G–I′) were transfected with a plasmid expressing GFP-SMNFL (A, D, and G), GFP-SMNΔ7 (B, E, and H), or GFP (C, F, and I). GFP-SMNFL is concentrated as aggregates in the cytoplasm and in the nucleus of neurons (A), fibroblasts (D), and HeLa cells (G). GFP-SMNΔ7 is localized in both the nucleus and cytoplasm of fibroblasts (E) and HeLa cells (H). B: Despite normal growth and survival of GFP-SMNΔ7-transfected neurons, only a very weak signal can be seen in the cytoplasm. Nuclei are stained with 4,6-diamidino-2-phenylindole. A′–C′, D′–F′, and G′–I′ are merged images. Scale bars: 10 μm (A–C′, G–I′); 20 μm (D–F′).
Figure 3
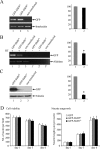
Characterization of primary cultures of cortical neurons transfected with a plasmid expressing GFP-SMNFL (lane 1), GFP-SMNΔ7 (lane 2), and nontransfected control (lane 3). Quantification of plasmid DNA through PCR amplification of GFP sequence (A), RNA using RT-PCR amplification (B), or GFP fusion protein by immunoblot experiment (C) 1 day after plating. Although a similar amount of transfected plasmid was used, a marked decrease in RNA (48%) and protein (81%) encoding GFP-SMNΔ7 was observed when compared with GFP-SMNFL. Relative amounts of plasmid DNA, RNA encoding GFP, and GFP protein were evaluated by determining the ratio of transgene to internal control (interleukin for DNA, aldolase for RNA, and tubulin for protein analyses). For transcript analysis, PCR amplification was performed with (+) or without (−) reverse transcriptase as a negative control. D: Cell viability was estimated from the mean number of nuclei on five same-phase contrast fields at 1, 3, and 6 days of culture. The neurite outgrowth was measured 1, 3, and 6 days after plating of transfected cortical neurons by a stereological procedure on five same-phase contrast fields. No statistically significant difference was observed whatever the transfected plasmids or the day of examination. Three independent experiments were performed. Bar means SE.
Figure 4
SMN protein stability assay in neurons and fibroblasts. Primary cultures of mouse cortical neurons (A) and embryonic fibroblasts (B) were transfected with a plasmid expressing GFP-SMNFL or GFP-SMNΔ7. Cells were treated with cycloheximide (CHX) 24 hours after transfection, and proteins were harvested at indicated time points (0, 1, 3, and 6 hours after CHX treatment). Similar amounts of total protein extracts were loaded and analyzed by Western blot using anti-GFP and anti-tubulin antibodies. C: Levels of GFP-SMNFL, GFP-SMNΔ7, and tubulin proteins were evaluated by densitometric analysis. Each point corresponds to the ratio of the fused protein: tubulin relative to that at time point 0.
Figure 5
Immunolabeling experiment of the 20S proteasome in HeLa and cortical neurons. Primary cultures of mouse cortical neurons (A–B‴) or HeLa cells (C–D‴) were transfected with a plasmid expressing GFP-SMNFL (A–A‴ and C–C‴) or GFP-SMNΔ7 (B–B‴ and D–D‴). The 20S proteasome is labeled in red (A′–D′) and GFP fluorescence is shown in green (A–D). Nuclei were stained with 4,6-diamidino-2-phenylindole (A‴–D‴). A″ to D″ are merged images. Scale bars: 10 μm (A-B‴); 20 μm (C–D‴).
Figure 6
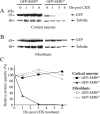
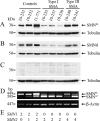
Correlation of SMNFL protein level with SMN2 gene copy number and the absence of SMNΔ7 protein in lymphoblastoid cell lines. Immunoblot analysis of proteins extracted from lymphoblastoid cell lines derived from controls, type I, or type III SMA patients. Immunoblots were incubated with hSMNex7-5381 (A), SMNtl monoclonal antibody (B), or with hSMNex8-5699 (C). Tubulin was used as internal control for loading. D: RT-PCR analysis of RNA extracted from the same cell lines, using primers flanking SMN exon 7, showing the absence of SMNΔ7 transcripts in the 10-253 cell line and the reduction of SMNFL transcripts in SMA patients. β-Actin was used as internal control. The 10-253 cell line, lacking the SMN2 gene, was used as a negative control for the detection of SMNΔ7 RNA or protein. E: The number of SMN1 or SMN2 genes in each control or patient is shown.
Figure 7
Absence of SMNΔ7 protein in fibroblasts of controls and SMA patients. Immunoblot analysis of total protein extracts from fibroblasts derived from controls or type I, II, or III SMA patients. A: The membranes were incubated with hSMNex8-5699 (top), SMNtl (middle), or tubulin (bottom). The 10-253 lymphoblastoid cell line was used as a negative control for the SMNΔ7 protein. HeLa cells transfected with the plasmid expressing GFP-SMNΔ7 (lane 2) were used as a positive control for the detection of SMNΔ7 protein when compared with GFP only (lane 1). B:SMN1 and SMN2 gene copy number in each control or patient. Asterisk indicates a nonspecific band because the 10-253 individual does not carry the SMN2 gene from which SMNΔ7 transcript is produced.
Figure 8
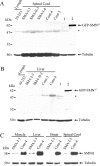
Absence of SMNΔ7 protein and dramatic reduction of SMNFL protein levels in SMA fetuses. Immunoblot analysis of proteins extracted from spinal cord (A) or liver (B). The membranes were incubated with hSMNex8-5699 antiserum specific to human SMNΔ7 protein. The 10-253 lymphoblastoid cell line (Lymph.) was used as a negative control and HeLa cells transfected with a plasmid expressing GFP-SMNΔ7 (lane 2) as a positive control when compared with GFP only (lane 1). C: Immunoblot analysis of proteins extracted from various tissues of the same type I SMA fetus compared with those of control fetus. Asterisk indicates a nonspecific band (see comment on Figure 7).
Similar articles
- A sensitive assay for measuring SMN mRNA levels in peripheral blood and in muscle samples of patients affected with spinal muscular atrophy.
Vezain M, Saugier-Veber P, Melki J, Toutain A, Bieth E, Husson M, Pedespan JM, Viollet L, Pénisson-Besnier I, Fehrenbach S, Bou J, Frébourg T, Tosi M. Vezain M, et al. Eur J Hum Genet. 2007 Oct;15(10):1054-62. doi: 10.1038/sj.ejhg.5201885. Epub 2007 Jul 4. Eur J Hum Genet. 2007. PMID: 17609673 - Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy.
Thurmond J, Butchbach ME, Palomo M, Pease B, Rao M, Bedell L, Keyvan M, Pai G, Mishra R, Haraldsson M, Andresson T, Bragason G, Thosteinsdottir M, Bjornsson JM, Coovert DD, Burghes AH, Gurney ME, Singh J. Thurmond J, et al. J Med Chem. 2008 Feb 14;51(3):449-69. doi: 10.1021/jm061475p. Epub 2008 Jan 19. J Med Chem. 2008. PMID: 18205293 - Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells.
Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. Grzeschik SM, et al. Ann Neurol. 2005 Aug;58(2):194-202. doi: 10.1002/ana.20548. Ann Neurol. 2005. PMID: 16049920 - Animal models of spinal muscular atrophy.
Schmid A, DiDonato CJ. Schmid A, et al. J Child Neurol. 2007 Aug;22(8):1004-12. doi: 10.1177/0883073807305667. J Child Neurol. 2007. PMID: 17761656 Review. - An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA).
Wirth B. Wirth B. Hum Mutat. 2000;15(3):228-37. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. Hum Mutat. 2000. PMID: 10679938 Review.
Cited by
- Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle.
Piazzon N, Schlotter F, Lefebvre S, Dodré M, Méreau A, Soret J, Besse A, Barkats M, Bordonné R, Branlant C, Massenet S. Piazzon N, et al. Nucleic Acids Res. 2013 Jan;41(2):1255-72. doi: 10.1093/nar/gks1224. Epub 2012 Dec 5. Nucleic Acids Res. 2013. PMID: 23221635 Free PMC article. - Skeletal muscle DNA damage precedes spinal motor neuron DNA damage in a mouse model of Spinal Muscular Atrophy (SMA).
Fayzullina S, Martin LJ. Fayzullina S, et al. PLoS One. 2014 Mar 25;9(3):e93329. doi: 10.1371/journal.pone.0093329. eCollection 2014. PLoS One. 2014. PMID: 24667816 Free PMC article. - Spinal muscular atrophy.
D'Amico A, Mercuri E, Tiziano FD, Bertini E. D'Amico A, et al. Orphanet J Rare Dis. 2011 Nov 2;6:71. doi: 10.1186/1750-1172-6-71. Orphanet J Rare Dis. 2011. PMID: 22047105 Free PMC article. Review. - The role of survival motor neuron protein (SMN) in protein homeostasis.
Chaytow H, Huang YT, Gillingwater TH, Faller KME. Chaytow H, et al. Cell Mol Life Sci. 2018 Nov;75(21):3877-3894. doi: 10.1007/s00018-018-2849-1. Epub 2018 Jun 5. Cell Mol Life Sci. 2018. PMID: 29872871 Free PMC article. Review. - Spinal Muscular Atrophy Therapeutics: Where do we Stand?
d'Ydewalle C, Sumner CJ. d'Ydewalle C, et al. Neurotherapeutics. 2015 Apr;12(2):303-16. doi: 10.1007/s13311-015-0337-y. Neurotherapeutics. 2015. PMID: 25631888 Free PMC article. Review.
References
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Frezal J, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. - PubMed
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical