A glycomics platform for the analysis of permethylated oligosaccharide alditols - PubMed (original) (raw)
A glycomics platform for the analysis of permethylated oligosaccharide alditols
Catherine E Costello et al. J Am Soc Mass Spectrom. 2007 Oct.
Abstract
This communication reports the development of an LC/MS platform for the analysis of permethylated oligosaccharide alditols that, for the first time, demonstrates routine online oligosaccharide isomer separation of these compounds before introduction into the mass spectrometer. The method leverages a high-resolution liquid chromatography system with the superior fragmentation pattern characteristics of permethylated oligosaccharide alditols that are dissociated under low-energy collision conditions using quadrupole orthogonal time-of-flight (QoTOF) instrumentation and up to pseudo MS(3) mass spectrometry. Glycoforms, including isomers, are readily identified and their structures assigned. The isomer-specific spectra include highly informative cross-ring and elimination fragments, branch position specific signatures, and glycosidic bond fragments, thus facilitating linkage, branch, and sequence assignment. The method is sensitive and can be applied using as little as 40 fmol of derivatized oligosaccharide. Because permethylation renders oligosaccharides nearly chemically equivalent in the mass spectrometer, the method is semiquantitative and, in this regard, is comparable to methods reported using high field NMR and capillary electrophoresis. In this postgenomic age, the importance of glycosylation in biological processes has become clear. The nature of many of the important questions in glycomics is such that sample material is often extremely limited, thus necessitating the development of highly sensitive methods for rigorous structural assignment of the oligosaccharides in complex mixtures. The glycomics platform presented here fulfills these criteria and should lead to more facile glycomics analyses.
Figures
Figure 1
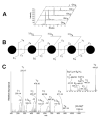
LC/MS analysis of permethylated maltooligosaccharide alditols. A, Extracted ion chromatograms of permethylated oligosaccharide alditol maltooligosaccharide series Glc4-8. The Glc4, Glc5 Glc6, Glc7 [M+Na]+ ions, Glc8 [M+2Na]2+ were detected at m/z 901.45, 1105.56, 1309.66, 1513.76 and 870.43 respectively. Integrated peak areas are proportional to abundances of the glycoforms. B and C, fragment origin and CID QoTOF MS/MS spectra of Glc5, [M+Na]+ m/z 1105.56. Fragmentation nomenclature is that of Domon and Costello . All fragments contain sodium. Monosaccharide component symbols are those suggested by Varki et al. Key ions are indicated. The inset in C demonstrates that distinct B, Z, C and Z ions are produced from the reduced and permethylated Hexose oligosaccharide.
Figure 2
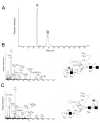
Extracted ion chromatogram of permethylated oligosaccharide alditols Man5-9GlcNAc2: A, a stacked plot of molecular ions for Man5-9GlcNAc2is shown; B, Three partially overlapping peaks are observed for Man7GlcNAc2 detected at [M+2Na]2+ m/z 1013.49 (bottom) and are labeled as D1, D2, and D3 and three are observed for Man8GlcNAc2 detected at [M+2Na]2+ m/z 1115.55 and are labeled as D1D2, D2D3 and D1D3 (top); C, Selected ion chromatogram of Man9GlcNAc2 produced from approximately 160 fmol. The molecular ion region of the mass spectrum is shown in the inset. See text for details concerning nomenclature.
Figure 3
CID QoTOF MS/MS spectra of Man7GlcNAc2 D1, D2, and D3 isomers. Major product ions, derived from [M+2Na]2+ m/z 1013.49 are indicated in each spectrum and their origins depicted to the structural representations to the right of each spectrum. Spectra and structural diagrams are shown in the order of their elution: A, Man7GlcNAc2 D2; B, Man7GlcNAc2 D3, and C, Man7GlcNAc2 D1. All fragments contain sodium.
Figure 4
LC/MS analysis of articular cartilage decorin _N_-glycans. A, The extracted ion chromatogram of articular cartilage decorin dHex1Hex4HexNAc4 permethylated oligosaccharide alditol is shown. Two well resolved peaks are observed for [M+2Na]2+ m/z 1039.56 and are labeled as A and B. B and C, CID QoTOF MS/MS spectra of articular cartilage decorin dHex1Hex4HexNAc4 permethylated oligosaccharide alditol isomers. The [M+2Na]2+ ions were isolated at m/z 1039.56. Key ions are indicated in each spectrum (A and C) and their origins are depicted in the structural representations to the right of each spectrum (B and C). All fragments contain sodium.
Figure 5
Trace ion chromatogram of reduced and permethylated C. elegans Hex3HexNAc1. Three major peaks are observed for [M+Na]+ m/z 942.55 and are labeled as A, B and C.
Figure 6
CID QoTOF MS/MS spectra of C. elegans Hex3GalNAc1 permethylated oligosaccharide alditol isomers isolated at [M+Na]+ m/z 942.55. The spectra of Peaks A, B and C are shown. Key ions are indicated in each spectrum and their origins and resultant linkage assignments are depicted in the structural representations to the right of each spectrum. Peaks B and C each contained two isomers, the lower arm of which was either Hex1,4Hex1,3- or Hex1,6Hex1,3-linked to the core GalNAc-ol. Specific structural assignments are based on pseudo MS3 spectrum shown in Figure 9. All fragments contain sodium. Monosaccharide identities are based on those of Varkii et al., [49] as in other figures except that hexoses that can be either Gal or Glc are represented as circles with an X.
Figure 7
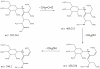
Pseudo MS3 spectra of key product ions of Hex3GalNAc1 permethylated alditol isomers. A:3X2 m/z 415.19 ion of Peak A, B; Z1α m/z 502.26 ion for Peak B, C; Z1α m/z 502.26 ion for Peak C, and D; 1,4A2α m/z 463.22 ion of Peak C. Proposed structures are shown above their spectra. In B and C, the Z1α m/z 502.26 ion can be either a 1,4 or 1,6 glycosidic bond. See text for details. All fragments contain sodium.
Scheme I
Decomposition of the 1-4-linked disaccharide isolated at m/z 502.26. The proposed pathway for the formation of key fragment ions is shown.
Similar articles
- Structural analysis of permethylated oligosaccharides using electrospray ionization quadrupole time-of-flight tandem mass spectrometry and deutero-reduction.
Morelle W, Faid V, Michalski JC. Morelle W, et al. Rapid Commun Mass Spectrom. 2004;18(20):2451-64. doi: 10.1002/rcm.1640. Rapid Commun Mass Spectrom. 2004. PMID: 15384134 - Fragmentation characteristics of permethylated oligosaccharides using a matrix-assisted laser desorption/ionization two-stage time-of-flight (TOF/TOF) tandem mass spectrometer.
Morelle W, Slomianny MC, Diemer H, Schaeffer C, van Dorsselaer A, Michalski JC. Morelle W, et al. Rapid Commun Mass Spectrom. 2004;18(22):2637-49. doi: 10.1002/rcm.1668. Rapid Commun Mass Spectrom. 2004. PMID: 15481102 - Methods in enzymology: O-glycosylation of proteins.
Peter-Katalinić J. Peter-Katalinić J. Methods Enzymol. 2005;405:139-71. doi: 10.1016/S0076-6879(05)05007-X. Methods Enzymol. 2005. PMID: 16413314 Review. - Analysis of methylated O-glycan alditols by reversed-phase NanoLC coupled CAD-ESI mass spectrometry.
Hanisch FG, Müller S. Hanisch FG, et al. Methods Mol Biol. 2009;534:107-15. doi: 10.1007/978-1-59745-022-5_8. Methods Mol Biol. 2009. PMID: 19277532 Review.
Cited by
- New strategies for profiling and characterization of human milk oligosaccharides.
Porfirio S, Archer-Hartmann S, Moreau GB, Ramakrishnan G, Haque R, Kirkpatrick BD, Petri WA, Azadi P. Porfirio S, et al. Glycobiology. 2020 Sep 28;30(10):774-786. doi: 10.1093/glycob/cwaa028. Glycobiology. 2020. PMID: 32248230 Free PMC article. - Glycoproteomics-based identification of cancer biomarkers.
Kim EH, Misek DE. Kim EH, et al. Int J Proteomics. 2011;2011:601937. doi: 10.1155/2011/601937. Epub 2011 Sep 28. Int J Proteomics. 2011. PMID: 22084691 Free PMC article. - Populations of metal-glycan structures influence MS fragmentation patterns.
Zhu F, Glover MS, Shi H, Trinidad JC, Clemmer DE. Zhu F, et al. J Am Soc Mass Spectrom. 2015 Jan;26(1):25-35. doi: 10.1007/s13361-014-1000-2. Epub 2014 Oct 15. J Am Soc Mass Spectrom. 2015. PMID: 25315458 Free PMC article. - Characterization of isomeric glycan structures by LC-MS/MS.
Veillon L, Huang Y, Peng W, Dong X, Cho BG, Mechref Y. Veillon L, et al. Electrophoresis. 2017 Sep;38(17):2100-2114. doi: 10.1002/elps.201700042. Epub 2017 May 17. Electrophoresis. 2017. PMID: 28370073 Free PMC article. Review. - Glycosaminoglycan Analysis by Cryogenic Messenger-Tagging IR Spectroscopy Combined with IMS-MS.
Khanal N, Masellis C, Kamrath MZ, Clemmer DE, Rizzo TR. Khanal N, et al. Anal Chem. 2017 Jul 18;89(14):7601-7606. doi: 10.1021/acs.analchem.7b01467. Epub 2017 Jul 7. Anal Chem. 2017. PMID: 28636333 Free PMC article.
References
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473(1):4–8. - PubMed
- Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14(2):95–101. - PubMed
- Gorelik E, Galili U, Raz A. On the role of cell surface carbohydrates and their binding proteins (lectins) in tumor metastasis. Cancer Metastasis Rev. 2001;20(3-4):245–77. - PubMed
- Lowe JB. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol. 2003;15(5):531–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources