Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases - PubMed (original) (raw)
Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases
Anton P Bussink et al. Genetics. 2007 Oct.
Abstract
Family 18 of glycosyl hydrolases encompasses chitinases and so-called chi-lectins lacking enzymatic activity due to amino acid substitutions in their active site. Both types of proteins widely occur in mammals although these organisms lack endogenous chitin. Their physiological function(s) as well as evolutionary relationships are still largely enigmatic. An overview of all family members is presented and their relationships are described. Molecular phylogenetic analyses suggest that both active chitinases (chitotriosidase and AMCase) result from an early gene duplication event. Further duplication events, followed by mutations leading to loss of chitinase activity, allowed evolution of the chi-lectins. The homologous genes encoding chitinase(-like) proteins are clustered in two distinct loci that display a high degree of synteny among mammals. Despite the shared chromosomal location and high homology, individual genes have evolved independently. Orthologs are more closely related than paralogues, and calculated substitution rate ratios indicate that protein-coding sequences underwent purifying selection. Substantial gene specialization has occurred in time, allowing for tissue-specific expression of pH optimized chitinases and chi-lectins. Finally, several family 18 chitinase-like proteins are present only in certain lineages of mammals, exemplifying recent evolutionary events in the chitinase protein family.
Figures
Figure 1.—
Protein alignment of the mature 39-kDa catalytic domain of mammalian chitinase(-like) proteins. Completely conserved amino acids are indicated a with solid baskground and those with a high similarity score (according to RISLER) are in a shaded background. The secondary structures (strands, arrows; helices and turns, T's) of human chitotriosidase, as published, are superimposed on the alignment. Cysteines involved in disulfide bridging are indicated by the numbers below the alignment. Chito, chitotriosidase; hcGP39, human cartilage glycoprotein 39; Ovi, oviductin; h, H. sapiens; m, M. musculus; r, R. norvegicus; b, B. taurus; c, Capra hircus; o, Ovis aries; s, Sus scrofa; cf, Canis familiaris; ma, Mesocricetus auratus. For further details, see text.
Figure 1.—
Protein alignment of the mature 39-kDa catalytic domain of mammalian chitinase(-like) proteins. Completely conserved amino acids are indicated a with solid baskground and those with a high similarity score (according to RISLER) are in a shaded background. The secondary structures (strands, arrows; helices and turns, T's) of human chitotriosidase, as published, are superimposed on the alignment. Cysteines involved in disulfide bridging are indicated by the numbers below the alignment. Chito, chitotriosidase; hcGP39, human cartilage glycoprotein 39; Ovi, oviductin; h, H. sapiens; m, M. musculus; r, R. norvegicus; b, B. taurus; c, Capra hircus; o, Ovis aries; s, Sus scrofa; cf, Canis familiaris; ma, Mesocricetus auratus. For further details, see text.
Figure 1.—
Protein alignment of the mature 39-kDa catalytic domain of mammalian chitinase(-like) proteins. Completely conserved amino acids are indicated a with solid baskground and those with a high similarity score (according to RISLER) are in a shaded background. The secondary structures (strands, arrows; helices and turns, T's) of human chitotriosidase, as published, are superimposed on the alignment. Cysteines involved in disulfide bridging are indicated by the numbers below the alignment. Chito, chitotriosidase; hcGP39, human cartilage glycoprotein 39; Ovi, oviductin; h, H. sapiens; m, M. musculus; r, R. norvegicus; b, B. taurus; c, Capra hircus; o, Ovis aries; s, Sus scrofa; cf, Canis familiaris; ma, Mesocricetus auratus. For further details, see text.
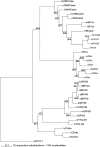
Figure 2.—
Maximum-likelihood tree of mammalian chitinase(-like) genes. The number on every node indicates the support bootstrap value for the likelihood analyses of a thousand replicate data sets. Only values other than 1000 are shown. The tree was rerooted by an outgroup (ceCht-1, Caenorhabditis elegans chitinase-1). Abbreviations as in Figure 1. For further details, see text.
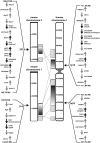
Figure 3.—
Synteny of loci encoding chitinase(-like) proteins between mice and humans. Schematic overview of the synteny of mouse locus 1F4 with human 1q32 and mouse locus 3F3 with human 1p13. The orientation and position of the genes are indicated with arrows. The genes of members of the chitinase protein family are depicted by solid arrows, whereas the other genes in the loci are depicted by shaded arrows.
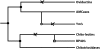
Figure 4.—
Overview of the evolution of chitinase(-like) genes. •, the “ancestral” gene duplications; ○, rodent-specific gene duplication; □, artiodactyle-specific gene duplication; a cross indicates the loss of catalytic activity mutations. “Chito-lectins” are chi-lectins evolved from the chitotriosidase gene (duplication).
Similar articles
- Chitinases and immunity: Ancestral molecules with new functions.
Di Rosa M, Distefano G, Zorena K, Malaguarnera L. Di Rosa M, et al. Immunobiology. 2016 Mar;221(3):399-411. doi: 10.1016/j.imbio.2015.11.014. Epub 2015 Dec 7. Immunobiology. 2016. PMID: 26686909 Review. - New paralogues and revised time line in the expansion of the vertebrate GH18 family.
Hussain M, Wilson JB. Hussain M, et al. J Mol Evol. 2013 Apr;76(4):240-60. doi: 10.1007/s00239-013-9553-4. Epub 2013 Apr 5. J Mol Evol. 2013. PMID: 23558346 - Molecular cloning and functional characterization of mouse chitotriosidase.
Zheng T, Rabach M, Chen NY, Rabach L, Hu X, Elias JA, Zhu Z. Zheng T, et al. Gene. 2005 Aug 29;357(1):37-46. doi: 10.1016/j.gene.2005.05.006. Gene. 2005. PMID: 16005164 - Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family.
Funkhouser JD, Aronson NN Jr. Funkhouser JD, et al. BMC Evol Biol. 2007 Jun 26;7:96. doi: 10.1186/1471-2148-7-96. BMC Evol Biol. 2007. PMID: 17594485 Free PMC article. - From bacteria to human: a journey into the world of chitinases.
Adrangi S, Faramarzi MA. Adrangi S, et al. Biotechnol Adv. 2013 Dec;31(8):1786-95. doi: 10.1016/j.biotechadv.2013.09.012. Epub 2013 Oct 3. Biotechnol Adv. 2013. PMID: 24095741 Review.
Cited by
- YKL-40 is differentially expressed in human embryonic stem cells and in cell progeny of the three germ layers.
Brøchner CB, Johansen JS, Larsen LA, Bak M, Mikkelsen HB, Byskov AG, Andersen CY, Møllgård K. Brøchner CB, et al. J Histochem Cytochem. 2012 Mar;60(3):188-204. doi: 10.1369/0022155411433331. Epub 2011 Dec 1. J Histochem Cytochem. 2012. PMID: 22140133 Free PMC article. - Significance of chitinase-3-like protein 1 in the pathogenesis of inflammatory diseases and cancer.
Yu JE, Yeo IJ, Han SB, Yun J, Kim B, Yong YJ, Lim YS, Kim TH, Son DJ, Hong JT. Yu JE, et al. Exp Mol Med. 2024 Feb;56(1):1-18. doi: 10.1038/s12276-023-01131-9. Epub 2024 Jan 4. Exp Mol Med. 2024. PMID: 38177294 Free PMC article. Review. - Targeting AMCase reduces esophageal eosinophilic inflammation and remodeling in a mouse model of egg induced eosinophilic esophagitis.
Cho JY, Rosenthal P, Miller M, Pham A, Aceves S, Sakuda S, Broide DH. Cho JY, et al. Int Immunopharmacol. 2014 Jan;18(1):35-42. doi: 10.1016/j.intimp.2013.10.026. Epub 2013 Nov 13. Int Immunopharmacol. 2014. PMID: 24239745 Free PMC article. - Innate Immunity of the Lung: From Basic Mechanisms to Translational Medicine.
Hartl D, Tirouvanziam R, Laval J, Greene CM, Habiel D, Sharma L, Yildirim AÖ, Dela Cruz CS, Hogaboam CM. Hartl D, et al. J Innate Immun. 2018;10(5-6):487-501. doi: 10.1159/000487057. Epub 2018 Feb 13. J Innate Immun. 2018. PMID: 29439264 Free PMC article. Review. - The Role of Glycoside Hydrolases in Phytopathogenic Fungi and Oomycetes Virulence.
Rafiei V, Vélëz H, Tzelepis G. Rafiei V, et al. Int J Mol Sci. 2021 Aug 28;22(17):9359. doi: 10.3390/ijms22179359. Int J Mol Sci. 2021. PMID: 34502268 Free PMC article. Review.
References
- Badariotti, F., M. Kypriotou, C. Lelong, M. P. Dubos, E. Renard et al., 2006. The phylogenetically conserved molluscan chitinase-like protein 1 (Cg-Clp1), homologue of human HC-gp39, stimulates proliferation and regulates synthesis of extracellular matrix components of mammalian chondrocytes. J. Biol. Chem. 281: 29583–29596. - PubMed
- Beutler, E., and G. A. Grabowski, 1995. Gaucher disease, pp. 2641–2670 in The Metabolic and Molecular Bases of Inherited Disease, Ed. 7, edited by C. R. Scriver, A. L. Beaudet, W. S. Sly and D. Valle. McGraw-Hill, New York.
- Boot, R. G., G. H. Renkema, A. Strijland, A. J. van Zonneveld and J. M. Aerts, 1995. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 270: 26252–26256. - PubMed
- Boot, R. G., G. H. Renkema, M. Verhoek, A. Strijland, J. Bliek et al., 1998. The human chitotriosidase gene. Nature of inherited enzyme deficiency. J. Biol. Chem. 273: 25680–25685. - PubMed
- Boot, R. G., E. F. Blommaart, E. Swart, K. Ghauharali-van der Vlugt, N. Bijl et al., 2001. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 276: 6770–6778. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases