Functional association of Sun1 with nuclear pore complexes - PubMed (original) (raw)
Functional association of Sun1 with nuclear pore complexes
Qian Liu et al. J Cell Biol. 2007.
Abstract
Sun1 and 2 are A-type lamin-binding proteins that, in association with nesprins, form a link between the inner nuclear membranes (INMs) and outer nuclear membranes of mammalian nuclear envelopes. Both immunofluorescence and immunoelectron microscopy reveal that Sun1 but not Sun2 is intimately associated with nuclear pore complexes (NPCs). Topological analyses indicate that Sun1 is a type II integral protein of the INM. Localization of Sun1 to the INM is defined by at least two discrete regions within its nucleoplasmic domain. However, association with NPCs is dependent on the synergy of both nucleoplasmic and lumenal domains. Cells that are either depleted of Sun1 by RNA interference or that overexpress dominant-negative Sun1 fragments exhibit clustering of NPCs. The implication is that Sun1 represents an important determinant of NPC distribution across the nuclear surface.
Figures
Figure 1.
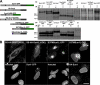
The mammalian SUN protein family. Sun1 features four hydrophobic sequences, H1–H4, each of roughly 20 amino acid residues. Its membrane-spanning domain is contained within the H2–H4 region. The Sun1 N terminus, including H1, is nucleoplasmic. The C-terminal SUN domain resides in the PNS. Murine but not primate Sun1 contains a predicted C2H2 zinc finger. Several splice isoforms of mouse Sun1 have been identified that feature the loss of sequences encoded by exons 6–8, including H1. Corresponding GenBank/EMBL/DDBJ accession nos. are BAB29445 (Sun1Δ6–8), AAT90501 (Sun1Δ6), and BAC29339 (Sun1Δ8). Four other mammalian SUN proteins are known. Sun2 is ubiquitously expressed and localizes to the INM. Sun3 (SUNC1), Sun4 (SPAG4), and Sun5 (SPAG4L; GenBank/EMBL/DDBJ accession no. NP_542406) appear to be expressed primarily in testis (unpublished data). When expressed in HeLa cells, Sun3 localizes to the NE (Fig. S2, available at
http://www.jcb.org/cgi/content/full/jcb.200704108/DC1
), whereas Sun4 (Hasan et al., 2006) and Sun5 (unpublished data) localize primarily to the ER.
Figure 2.
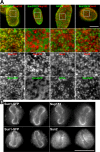
Sun1 and 2 are segregated within the plane of the NE. Immunofluorescence microscopy of HeLa cells stably expressing mouse Sun1-GFP or human Sun2-GFP using antinucleoporin and anti-SUN protein antibodies. (A) Images of the nuclear surface reveal Sun1-GFP colocalization with Nup153. In contrast, the more diffuse Sun2-GFP is found in NPC-free regions. Endogenous Sun2 displays no colocalization with either NPCs labeled with the antinucleoporin antibody QE5 or with Sun1-GFP. The boxed areas in the top panels are magnified in the bottom three panels. (B) At late anaphase to early telophase, reforming nuclei exhibit a distinct distribution of NPC and NE components. Sun1-GFP localizes with Nup153 at the lateral margins of the mass of newly segregated chromatids and is absent from the Sun2-positive core region. These data suggest that Sun1 is closely associated with NPCs, a pattern that is established early in NE formation. Bars (A), 5 μm; (B) 4 μm.
Figure 3.
Sun1 but not Sun2 is closely associated with NPCs as revealed by immunoelectron microscopy. (A) Views of NPC cross sections from tetracycline-induced HeLa cells expressing Sun1-GFP that reveal anti-GFP–associated gold particles close to NPCs. (B) Sections of uninduced cells display little or no gold labeling. (C) Images of NE cross sections from HeLa cells expressing Sun2-GFP reveal gold particles in the PNS but with no preferential association with NPCs. c, cytoplasm; n, nucleus. Arrowheads indicate gold particles. (D and E) Quantitative analysis of the distribution of gold particles from NE cross sections of HeLa cells expressing Sun1- (D) or Sun2-GFP (E). The position of gold particles, which is defined by horizontal distance (from the NPC eightfold axis) and vertical distance (from the central plane of the NE), was measured in cross sectioned NEs (as in A and C) and plotted in a single dot graphic. Micrographs are provided as a visual reference for the position of the gold particles. Histograms for the distribution of gold particles for horizontal and vertical distances are shown on adjacent panels. For both Sun1 and 2, gold particles were scored within 200- and 600-nm windows on either side of each NPC. The larger, more conservative window size still reveals an absolute eightfold higher labeling density of Sun1 over Sun2 within 120 nm of the NPC center, with a peak density at 66 nm. A total of 88 and 92 gold particles were scored for D and E, respectively. Because of the far broader distribution of Sun2-GFP, the scale in E is five times that in D. Bars, 100 nm.
Figure 4.
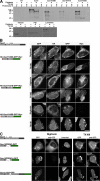
Sun1 contains a single TM domain. (A) Three Sun1 mutants containing the N-terminal domain followed by H2, H2–H3, and H2–H4 were tagged with HA at the N terminus and GFP followed by a myc epitope at the C terminus. HeLa cells transfected (or not transfected [NT]) with these constructs were labeled with [35S]Met/Cys, permeabilized with digitonin, and incubated with proteinase K. After SDS lysis, immunoprecipitation with anti-myc, and SDS-PAGE analysis, only Sun1N455 retained a protected fragment of the predicted size for the GFP (∼30 kD). Permeabilization with TX-100 resulted in complete protein degradation. Nontransfected cells served as a negative control, whereas Sun1-GFP provided a positive control, with a 65–70-kD protected fragment. Western blot analysis was used to confirm the effectiveness of the digitonin permeabilization. Tubulin and lamins A/C were degraded after either digitonin or TX-100 permeabilization. In contrast, the ER lumenal protein PDI remained intact after digitonin permeabilization but was degraded after TX-100 treatment. (B) To further establish the orientation of these Sun1 constructs, 24 h after transfection, the HeLa cells were fixed and permeabilized with either digitonin or TX-100. Analyses focused on cells expressing sufficiently high levels of recombinant protein such that GFP fluorescence could be observed in both the NE and ER/cytoplasmic membranes. With digitonin permeabilization, both myc and HA epitopes were readily detected for HA-Sun1N380-GFP-myc and HA-Sun1N415-GFP-myc. In contrast, the HA but not C-terminal myc epitope tag was accessible for HA-Sun1N455-GFP-myc. In all cases, both myc and HA tags were accessible after TX-100 permeabilization. (C) Three more C-terminal GFP-tagged Sun1 constructs containing the first 220 residues of Sun1 fused to H3 or H3–H4 as well as full-length Sun1 lacking H2–H3 (Sun1N220H3-GFP, Sun1N220H34-GFP, and Sun1ΔH23-GFP, respectively) were permeabilized as described in A and labeled with anti-GFP antibodies. With digitonin permeabilization, the presence of the H4 domain rendered the GFP moiety inaccessible to antibody. Collectively, these results indicate that H4 serves as Sun1's sole TM domain. Bars, 4 μm.
Figure 5.
The Sun1 nucleoplasmic domain has overlapping NE localization motifs. Immunofluorescence microscopy of HeLa cells transiently expressing Sun1 deletion constructs. (A) Sun1N355, a region previously identified as conferring NE localization, was further mutated based on a natural splice isoform of Sun1 (Sun1N355Δ221–343-GFP). This protein, which lacks the H1 domain, is predominantly nucleoplasmic. Upon the inclusion of H2 (Sun1N380Δ221–343-GFP), it becomes NE associated. Deletion of the N-terminal 220 residues from the NE-localized Sun1N380 (myc-Sun1-221–380) fails to eliminate NE localization. However, extension of the deletion to include H1 (myc-Sun1-261–380) results in an ER localization with little concentration in the NE. (B) To examine the role of H2–H4 in the localization of Sun1, H234-GFP was expressed in HeLa cells, where it concentrates in the Golgi apparatus. This region (H2–H4) of Sun1 was replaced with the TM domain of Sun3 (HA-Sun1(S3TM)), resulting in localization that was indistinguishable from full-length Sun1. However, upon deletion of the nucleoplasmic domain (S3TMSun1L-GFP), the protein was found largely in the Golgi apparatus. Similarly, deletion of the H2 and H3 domain from H234Sun1L (H4Sun1L-GFP) led to the loss of NE association with a predominantly ER localization. NE localization of Golgi-associated S3TMSun1L-GFP could be partially rescued by addition of the H2–H3 sequence (H23(S3TM)Sun1L-GFP) to the N terminus. These data identify two overlapping regions of the Sun1 nucleoplasmic domain that are sufficient for NE targeting, H1–H2 and H2–H3. However, the latter is only functional in the context of a TM sequence and the Sun1 lumenal domain. Bars, 5 μm.
Figure 6.
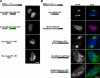
Sun1 forms homotypic oligomers in vivo. (A) To define the oligomerization state of Sun, HA-Sun1 was cotransfected into HeLa cells with Sun1-GFP, Sun1N455-GFP, H234Sun1L-GFP, and S3TMSun1L-GFP. All samples were labeled with [35S]Met/Cys and immunoprecipitated with anti-GFP antibodies. Full-length HA-Sun1 was most efficiently coprecipitated with Sun1-GFP followed by the lumenal then nucleoplasmic domain containing fusion proteins. NT, nontransfected. (B) In vivo evidence of Sun1 oligomerization mediated by the lumenal domain was provided by the cotransfection of Sun1-GFP or HA-Sun1 with SS-HA-Sun1L-KDEL or S3TMSun1L-GFP, respectively. Alone, these proteins localize predominantly to the ER or Golgi apparatus, respectively. Coexpression of full-length Sun1 recruited both proteins to the NE. Thus, Sun1 forms homotypic oligomers that independently involve the lumenal and, to a lesser extent, the nucleoplasmic domains. Bar, 6 μm.
Figure 7.
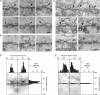
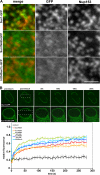
Sun1 association with NPCs requires both the nucleoplasmic and lumenal domains. (A) To define regions of Sun1 involved in NPC association, Sun1 mutants lacking either the lumenal (Sun1N455-GFP) or nucleoplasmic domains (H234Sun1L-GFP) were transiently expressed in HeLa cells. Neither Sun1 deletion mutant displayed obvious colocalization with Nup153. (B) To further analyze the roles of various domains in Sun1 targeting and retention, FRAP analysis was performed on Sun1, Sun1N455, Sun1N380, Sun1N380Δ221–343, Sun1N380Δ221–355, and H234Sun1L, each bearing a GFP tag at the C terminus. In contrast to full-length Sun1-GFP, which is relatively immobile in the NE, fluorescence recovery occurred for all deletion proteins within a span of ∼1 min. Together, these data indicate that it is the combination of the nucleoplasmic and lumenal domains that stabilizes Sun1 in the NE, potentially involving an association with NPCs. Dotted boxes define the photobleached region. Bar, 1 μm.
Figure 8.
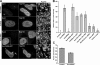
Perturbation of Sun1 affects NPC distribution. (A) Immunofluorescence microscopy of HeLa cells 48 h after SUN protein depletion by RNAi. Cells were double labeled with antibodies against Sun1 or Sun2 and Nup153. The loss of Sun1 was associated with altered nuclear morphology and changes in the distribution of NPCs. Magnification of the nuclear surface reveals large pore-free tracts between clusters of NPCs. Nontransfected cells (control for Sun1 and insets for Sun2) or Sun2 RNAi had no such effect. The expression of Sun1N455-GFP altered NPC distribution in a manner similar to Sun1 RNAi. Boxed areas are magnified in the right panels. (B) To quantify the observed changes in NPC distribution, the relative SD of binned pixel intensity of anti-Nup153 fluorescence intensity across the projected nuclear surface was calculated. All measurements were standardized relative to the nontransfected control, which was set at 0%. Sun1 RNAi and Sun1N455-GFP were most effective at inducing NPC clustering followed by H234Sun1L-GFP, myc-Sun1N220, and myc-Sun1 220–380. SS-HA-Sun1L-KDEL and Sun1-GFP had a minimal effect on NPC distribution. Lamin C served as a transfection control and induced no obvious NPC clustering. n = 7–18. (C) To quantify the altered nuclear morphology induced by Sun1 RNAi, the ratio of the projected nuclear area to the perimeter was measured. RNAi of Sun1 led to a 35% reduction in this ratio over control cells, which were set to 100%. n = 10–11. Error bars represent SEM. Bar, 5 μm.
Figure 9.
Sun1 topology and interactions. (A) Sun1 is envisaged as forming homodimers via interactions involving the membrane-proximal coiled coil within its C-terminal lumenal domain. Nucleoplasmic domain interactions may also contribute to homodimer formation. Sun1 functions as a tether for ONM nesprin proteins. Nesprins 1 and 2 provide links to the actin cytoskeleton, whereas nesprin 3 binds plectin, a versatile cytolinker. (B) Sun1 is associated with NPCs, and, in addition, its nucleoplasmic domain displays preferential binding to newly synthesized pre–lamin A. Therefore, Sun1 may provide a link between NPCs and the A-type lamins. In this way, the Sun1-mediated nucleation of A-type lamina assembly may occur at NPCs.
Similar articles
- The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope.
Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. Padmakumar VC, et al. J Cell Sci. 2005 Aug 1;118(Pt 15):3419-30. doi: 10.1242/jcs.02471. J Cell Sci. 2005. PMID: 16079285 - Effects of Inner Nuclear Membrane Proteins SUN1/UNC-84A and SUN2/UNC-84B on the Early Steps of HIV-1 Infection.
Schaller T, Bulli L, Pollpeter D, Betancor G, Kutzner J, Apolonia L, Herold N, Burk R, Malim MH. Schaller T, et al. J Virol. 2017 Sep 12;91(19):e00463-17. doi: 10.1128/JVI.00463-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28747499 Free PMC article. - Sun1 forms immobile macromolecular assemblies at the nuclear envelope.
Lu W, Gotzmann J, Sironi L, Jaeger VM, Schneider M, Lüke Y, Uhlén M, Szigyarto CA, Brachner A, Ellenberg J, Foisner R, Noegel AA, Karakesisoglou I. Lu W, et al. Biochim Biophys Acta. 2008 Dec;1783(12):2415-26. doi: 10.1016/j.bbamcr.2008.09.001. Epub 2008 Sep 19. Biochim Biophys Acta. 2008. PMID: 18845190 - Aspects of nuclear envelope dynamics in mitotic cells.
Burke B, Shanahan C, Salina D, Crisp M. Burke B, et al. Novartis Found Symp. 2005;264:22-30; discussion 30-4, 227-30. Novartis Found Symp. 2005. PMID: 15773745 Review. - Isolation and fractionation of rat liver nuclear envelopes and nuclear pore complexes.
Matunis MJ. Matunis MJ. Methods. 2006 Aug;39(4):277-83. doi: 10.1016/j.ymeth.2006.06.003. Methods. 2006. PMID: 16870471 Review.
Cited by
- The SUN2-nesprin-2 LINC complex and KIF20A function in the Golgi dispersal.
Hieda M, Matsumoto T, Isobe M, Kurono S, Yuka K, Kametaka S, Wang JY, Chi YH, Kameda K, Kimura H, Matsuura N, Matsuura S. Hieda M, et al. Sci Rep. 2021 Mar 8;11(1):5358. doi: 10.1038/s41598-021-84750-4. Sci Rep. 2021. PMID: 33686165 Free PMC article. - The scaffold nucleoporins SAR1 and SAR3 are essential for proper meiotic progression in Arabidopsis thaliana.
Fernández-Jiménez N, Martinez-Garcia M, Varas J, Gil-Dones F, Santos JL, Pradillo M. Fernández-Jiménez N, et al. Front Cell Dev Biol. 2023 Dec 4;11:1285695. doi: 10.3389/fcell.2023.1285695. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38111849 Free PMC article. - The LINC complex, mechanotransduction, and mesenchymal stem cell function and fate.
Bouzid T, Kim E, Riehl BD, Esfahani AM, Rosenbohm J, Yang R, Duan B, Lim JY. Bouzid T, et al. J Biol Eng. 2019 Aug 7;13:68. doi: 10.1186/s13036-019-0197-9. eCollection 2019. J Biol Eng. 2019. PMID: 31406505 Free PMC article. Review. - Chain reaction: LINC complexes and nuclear positioning.
Burke B. Burke B. F1000Res. 2019 Jan 31;8:F1000 Faculty Rev-136. doi: 10.12688/f1000research.16877.1. eCollection 2019. F1000Res. 2019. PMID: 30774932 Free PMC article. Review. - Molecular Insights into the Mechanisms of SUN1 Oligomerization in the Nuclear Envelope.
Jahed Z, Fadavi D, Vu UT, Asgari E, Luxton GWG, Mofrad MRK. Jahed Z, et al. Biophys J. 2018 Mar 13;114(5):1190-1203. doi: 10.1016/j.bpj.2018.01.015. Biophys J. 2018. PMID: 29539404 Free PMC article.
References
- Apel, E.D., R.M. Lewis, R.M. Grady, and J.R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986–31995. - PubMed
- Bodoor, K., S. Shaikh, D. Salina, W.H. Raharjo, R. Bastos, M. Lohka, and B. Burke. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264. - PubMed
- Broers, J.L., E.A. Peeters, H.J. Kuijpers, J. Endert, C.V. Bouten, C.W. Oomens, F.P. Baaijens, and F.C. Ramaekers. 2004. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous