Two human ARFGAPs associated with COP-I-coated vesicles - PubMed (original) (raw)
Two human ARFGAPs associated with COP-I-coated vesicles
Gabriella Frigerio et al. Traffic. 2007 Nov.
Abstract
ADP-ribosylation factors (ARFs) are critical regulators of vesicular trafficking pathways and act at multiple intracellular sites. ADP-ribosylation factor-GTPase-activating proteins (ARFGAPs) are proposed to contribute to site-specific regulation. In yeast, two distinct proteins, Glo3p and Gcs1p, together provide overlapping, essential ARFGAP function required for coat protein (COP)-I-dependent trafficking. In mammalian cells, only the Gcs1p orthologue, named ARFGAP1, has been characterized in detail. However, Glo3p is known to make the stronger contribution to COP I traffic in yeast. Here, based on a conserved signature motif close to the carboxy terminus, we identify ARFGAP2 and ARFGAP3 as the human orthologues of yeast Glo3p. By immunofluorescence (IF), ARFGAP2 and ARFGAP3 are closely colocalized with coatomer subunits in NRK cells in the Golgi complex and peripheral punctate structures. In contrast to ARFGAP1, both ARFGAP2 and ARFGAP3 are associated with COP-I-coated vesicles generated from Golgi membranes in the presence of GTP-gamma-S in vitro. ARFGAP2 lacking its zinc finger domain directly binds to coatomer. Expression of this truncated mutant (DeltaN-ARFGAP2) inhibits COP-I-dependent Golgi-to-endoplasmic reticulum transport of cholera toxin (CTX-K63) in vivo. Silencing of ARFGAP1 or a combination of ARFGAP2 and ARFGAP3 in HeLa cells does not decrease cell viability. However, silencing all three ARFGAPs causes cell death. Our data provide strong evidence that ARFGAP2 and ARFGAP3 function in COP I traffic.
Figures
Figure 1
ARFGAP domain and conserved motif of a representative subset of Glo3 proteins. A) The N-terminal ARFGAP domains of Glo3 and Gcs1 proteins from Saccharomyces cerevisiae, Schizosaccharomyces pombe and Homo sapiens were aligned using the
clustal
V algorithm in the M
eg
A
lign
program (DNAS
tar
package). Note the conserved cysteine residues highlighted in yellow and the overall high degree of sequence conservation in this domain. B). The Glo3 motif (highlighted in orange) consists of a repeat of 15 residues separated by 11–17 residues. It is a signature motif that allows unambiguous identification of Glo3 orthologues. Genbank accession numbers: cerevisiae: 6320969 (S. cerevisiae); Plasmodium 23478251 (P. yoelii); worm: 25153991 (Caenorhabditis elegans); fly: 24668642 (Drosophila melanogaster); plant A: 7487780, plant B: 18403775, plant C 15237500 (Arabidopsis thaliana); fish: assembled from several ESTs: 12148139, 12171549, 12158629 and 12265392 (Danio rerio); frog: 27695479 (Xenopus laevis) and human A: 21263420 and human B: 31543983 (H. sapiens).
Figure 2
Sequence alignment of yeast Glo3p and human ARFGAP2 and ARFGAP3. ARFGAP2 and ARFGAP3 share 49.6% protein sequence identity. ARFGAP2 and ARFGAP3 share 17.6 and 19.9% sequence identity with Glo3p, respectively.
Figure 3
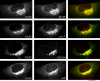
ARFGAP2 colocalizes with coatomer in NRK cells. Methanol–acetone-fixed NRK cells were stained with affinity-purified ARFGAP2 antibodies and monoclonal antibodies against β-COP, p115, GM130 or γ-adaptin. Cy3-coupled anti-rabbit and Alexa488-coupled anti-mouse secondary antibodies were used. Images were acquired using a Zeiss Axioplan Fluorescent Microscope equipped with a charge-coupled device camera. The individual channels are shown in monochrome; the composite overlay is shown to the right (green channel, ARFGAP2; red channel, β-COP, p115, GM130 and γAP, respectively). Bar = 10 μm.
Figure 4
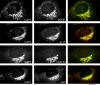
ARFGAP3 colocalizes with coatomer. Methanol–acetone fixed NRK cells were labelled with affinity-purified ARFGAP3 antibodies and mAbs against β-COP, p115, GM130 or γ-adaptin (see Figure 3 for other details). The individual channels are shown in monochrome; the composite overlay is shown to the right (green channel, ARFGAP3; red channel, β-COP, p115, GM130 and γAP, respectively). Bar = 10 μm.
Figure 5
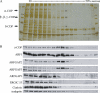
ARFGAP2 and ARFGAP3 are associated with COP I vesicles generated in vitro. Vesicle budding reactions were performed according to the protocol developed by , using the non-hydrolysable GTP analogue, GTP-γ-S. Vesicles produced in the reaction were separated from the Golgi donor membranes on sucrose density gradients and peaked at 40–43% sucrose. Fractions were analysed by silver stain (upper panel) and immunoblotting with antibodies as indicated (lower panel). The positions of coatomer bands are indicated on the left and also by asterisks. Note the abundant presence of ARFGAP2 and ARFGAP3 in the vesicle fractions defined by the presence of coatomer subunits (fractions 8 + 9) but the absence of ARFGAP1.
Figure 6
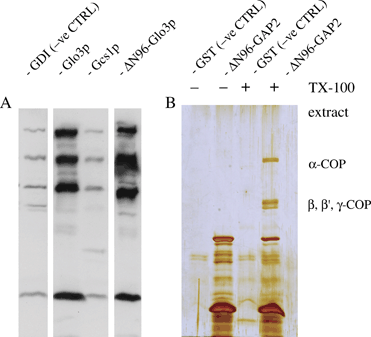
Direct interaction of yeast and human Glo3 with coatomer. His6- or GST-tagged proteins were used to pull down coatomer from yeast or rat liver cytosol, under conditions described by us previously . A) His6-tagged Glo3p lacking its amino terminus was compared with full-length Glo3p as well as full-length Gcs1p in its ability to bind coatomer from yeast cytosol. Protein bound to Ni-NTA agarose beads was resolved by SDS–PAGE, followed by Western blotting and incubation with anti-yeast coatomer antibodies detected by ECL. His-tagged Glo3p lacking its ARFGAP domain (ΔN96) still binds coatomer from yeast cytosol in vitro, whereas Gcs1p does not. GTP dissociation inhibitor is used as a negative control here. B) A GST-fusion protein harbouring ΔN96-ARFGAP2 was used for a pull-down from a centrifugation-cleared TX-100 extract of pig brain crude microsomal membranes. Glutathione S-transferase was used as the negative control. Note absence of binding in the negative control and the presence of the characteristic coatomer bands in the pull-down involving the ΔN96-ARFGAP2 GST fusion as the fishing hook. Enrichment of α- and β-COP in the bound fraction was confirmed using antipeptide antibodies (data not shown).
Figure 7
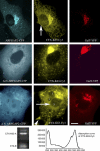
ARFGAP2 is involved in COP-I-dependent trafficking in Vero cells. Vero cells coexpressing full-length ARFGAP2–CFP and the Golgi marker GalT-YFP were treated with CTX-K63-Cy3 (upper panel). After 3 h of internalization, CTX-K63-Cy3, the Cy3-labelled A-subunit, is prominently present in fine ER structures (including the nuclear envelope; arrow) and in the Golgi complex. ARFGAP2–CFP colocalizes well with GalT-YFP in the Golgi. Additionally, ARFGAP2–CFP is present in scattered punctate structures, most likely intermediate compartment. In contrast, in Vero cells expressing ΔN-ARFGAP2–CFP2 transport of CTX-K63-Cy3 to the ER is inhibited (middle panel). ΔN-ARFGAP2–CFP2 localizes to the Golgi complex. Vero cells overexpressing ΔN-ARFGAP2–CFP2 for extended periods often display two to three nuclei and show severe inhibition of toxin transport (lower panel). The bottom panel is the characterization of the Cy3-labelled CTX-K63 (non-toxic AB5 holotoxin) used for the experiments (for details see methods). Mostly the A-subunit of CTX-K63 is labelled, with traces of B present. Scale bar: 10 μm.
Figure 8
Silencing of all three ARFGAPs is lethal in HeLa cells. Silencing of ARFGAP1 or a combination of ARFGAP2 and ARFGAP3 failed to cause lethality in HeLa cells, as well as in NRK cells (data not shown). A) The depletion of ARFGAP1, ARFGAP2 or ARFGAP3 in single knock-downs in HeLa cells was verified by immunoblotting. Antibodies against tubulin and γ-COP were used to verify equal protein loading. B and C) Cell counting was performed from DAPI-stained HeLa cells grown on coverslips. Note the strong reduction in cell numbers in the ARFGAP triple knock-down compared with a double knock-down involving ARFGAP2 and ARFGAP3 combined with a control oligo. For details of the oligos, see Materials and Methods. D) Quantification of the reduction in cell numbers from the above. More than 75% cell death was routinely observed after three sets of siRNA transfections at the 72-h time-point after the last transfection (in 10 independent experiments). Bar = 100 μm.
Similar articles
- ARFGAP2 and ARFGAP3 are essential for COPI coat assembly on the Golgi membrane of living cells.
Kartberg F, Asp L, Dejgaard SY, Smedh M, Fernandez-Rodriguez J, Nilsson T, Presley JF. Kartberg F, et al. J Biol Chem. 2010 Nov 19;285(47):36709-20. doi: 10.1074/jbc.M110.180380. Epub 2010 Sep 21. J Biol Chem. 2010. PMID: 20858901 Free PMC article. - Differential roles of ArfGAP1, ArfGAP2, and ArfGAP3 in COPI trafficking.
Weimer C, Beck R, Eckert P, Reckmann I, Moelleken J, Brügger B, Wieland F. Weimer C, et al. J Cell Biol. 2008 Nov 17;183(4):725-35. doi: 10.1083/jcb.200806140. J Cell Biol. 2008. PMID: 19015319 Free PMC article. - Three homologous ArfGAPs participate in coat protein I-mediated transport.
Saitoh A, Shin HW, Yamada A, Waguri S, Nakayama K. Saitoh A, et al. J Biol Chem. 2009 May 15;284(20):13948-13957. doi: 10.1074/jbc.M900749200. Epub 2009 Mar 19. J Biol Chem. 2009. PMID: 19299515 Free PMC article. - [Retrograde transport].
Aoe T. Aoe T. Tanpakushitsu Kakusan Koso. 2004 May;49(7 Suppl):914-5. Tanpakushitsu Kakusan Koso. 2004. PMID: 15168491 Review. Japanese. No abstract available. - ArfGAP1 function in COPI mediated membrane traffic: currently debated models and comparison to other coat-binding ArfGAPs.
Shiba Y, Randazzo PA. Shiba Y, et al. Histol Histopathol. 2012 Sep;27(9):1143-53. doi: 10.14670/HH-27.1143. Histol Histopathol. 2012. PMID: 22806901 Free PMC article. Review.
Cited by
- Arf GTPase-activating proteins and their potential role in cell migration and invasion.
Campa F, Randazzo PA. Campa F, et al. Cell Adh Migr. 2008 Oct-Dec;2(4):258-62. doi: 10.4161/cam.2.4.6959. Epub 2008 Oct 9. Cell Adh Migr. 2008. PMID: 19262159 Free PMC article. - COPI budding within the Golgi stack.
Popoff V, Adolf F, Brügger B, Wieland F. Popoff V, et al. Cold Spring Harb Perspect Biol. 2011 Nov 1;3(11):a005231. doi: 10.1101/cshperspect.a005231. Cold Spring Harb Perspect Biol. 2011. PMID: 21844168 Free PMC article. Review. - The Arf GAP Asap promotes Arf1 function at the Golgi for cleavage furrow biosynthesis in Drosophila.
Rodrigues FF, Shao W, Harris TJ. Rodrigues FF, et al. Mol Biol Cell. 2016 Oct 15;27(20):3143-3155. doi: 10.1091/mbc.E16-05-0272. Epub 2016 Aug 17. Mol Biol Cell. 2016. PMID: 27535433 Free PMC article. - Arf GAP2 is positively regulated by coatomer and cargo.
Luo R, Ha VL, Hayashi R, Randazzo PA. Luo R, et al. Cell Signal. 2009 Jul;21(7):1169-79. doi: 10.1016/j.cellsig.2009.03.006. Epub 2009 Mar 16. Cell Signal. 2009. PMID: 19296914 Free PMC article. - Mutational analysis of betaCOP (Sec26p) identifies an appendage domain critical for function.
DeRegis CJ, Rahl PB, Hoffman GR, Cerione RA, Collins RN. DeRegis CJ, et al. BMC Cell Biol. 2008 Jan 22;9:3. doi: 10.1186/1471-2121-9-3. BMC Cell Biol. 2008. PMID: 18211691 Free PMC article.
References
- Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. - PubMed
- Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nat Rev Mol Cell Biol. 2003;4:409–414. - PubMed
- Duden R. ER-to-Golgi transport: COP I & COP II function. Mol Membr Biol. 2003;20:197–207. - PubMed
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous