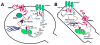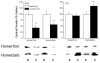Homers regulate drug-induced neuroplasticity: implications for addiction - PubMed (original) (raw)
Review
Homers regulate drug-induced neuroplasticity: implications for addiction
Karen K Szumlinski et al. Biochem Pharmacol. 2008.
Abstract
Drug addiction is a chronic, relapsing disorder, characterized by an uncontrollable motivation to seek and use drugs. Converging clinical and preclinical observations implicate pathologies within the corticolimbic glutamate system in the genetic predisposition to, and the development of, an addicted phenotype. Such observations pose cellular factors regulating glutamate transmission as likely molecular candidates in the etiology of addiction. Members of the Homer family of proteins regulate signal transduction through, and the trafficking of, glutamate receptors, as well as maintain and regulate extracellular glutamate levels in corticolimbic brain regions. This review summarizes the existing data implicating the Homer family of protein in acute behavioral and neurochemical sensitivity to drugs of abuse, the development of drug-induced neuroplasticity, as well as other behavioral and cognitive pathologies associated with an addicted state.
Figures
Figure 1
Illustration of the putative interactions between (A) constitutively expressed (CC) Homer proteins and (B) IEG Homer proteins with their EVH1-bound partners within the postsynaptic density of dendritic spines.
Figure 2
Homer protein levels are differentially regulated in the cell body and terminal regions of the corticoaccumbens glutamate pathway by cocaine. Immunoblotting for total Homer protein expression was conducted on NAC and PFC tissue from C57BL/6J mice at 3 weeks withdrawal from repeated cocaine (7 X 30 mg/kg) or saline administration. Bottom panels: Anti-Homer1b/c and anti-Homer2a/b primary antibodies recognized bands at 47 kDa [e.g., 39,144,169,295]. Left: Cocaine-treated mice showed an approximately 50% reduction in Homer1b/c (t20=2.34, p=0.03) and Homer2a/b (t20=2.32, p=0.03] within the NAC. Right: Cocaine-treated mice did not differ from saline controls regarding Homer1b/c expression (p=0.95), but exhibited an approximately 30% increase in Homer2a/b (t20=2.45, p=0.02). *p<0.05 for saline versus cocaine.
Similar articles
- Homer proteins regulate sensitivity to cocaine.
Szumlinski KK, Dehoff MH, Kang SH, Frys KA, Lominac KD, Klugmann M, Rohrer J, Griffin W 3rd, Toda S, Champtiaux NP, Berry T, Tu JC, Shealy SE, During MJ, Middaugh LD, Worley PF, Kalivas PW. Szumlinski KK, et al. Neuron. 2004 Aug 5;43(3):401-13. doi: 10.1016/j.neuron.2004.07.019. Neuron. 2004. PMID: 15294147 - Homer isoforms differentially regulate cocaine-induced neuroplasticity.
Szumlinski KK, Abernathy KE, Oleson EB, Klugmann M, Lominac KD, He DY, Ron D, During M, Kalivas PW. Szumlinski KK, et al. Neuropsychopharmacology. 2006 Apr;31(4):768-77. doi: 10.1038/sj.npp.1300890. Neuropsychopharmacology. 2006. PMID: 16160706 - Homer proteins: implications for neuropsychiatric disorders.
Szumlinski KK, Kalivas PW, Worley PF. Szumlinski KK, et al. Curr Opin Neurobiol. 2006 Jun;16(3):251-7. doi: 10.1016/j.conb.2006.05.002. Epub 2006 May 15. Curr Opin Neurobiol. 2006. PMID: 16704932 Review. - Neuroadaptations in the cellular and postsynaptic group 1 metabotropic glutamate receptor mGluR5 and Homer proteins following extinction of cocaine self-administration.
Ghasemzadeh MB, Vasudevan P, Mueller C, Seubert C, Mantsch JR. Ghasemzadeh MB, et al. Neurosci Lett. 2009 Mar 13;452(2):167-71. doi: 10.1016/j.neulet.2008.12.028. Epub 2008 Dec 24. Neurosci Lett. 2009. PMID: 19118598 Free PMC article. - New medications for drug addiction hiding in glutamatergic neuroplasticity.
Kalivas PW, Volkow ND. Kalivas PW, et al. Mol Psychiatry. 2011 Oct;16(10):974-86. doi: 10.1038/mp.2011.46. Epub 2011 Apr 26. Mol Psychiatry. 2011. PMID: 21519339 Free PMC article. Review.
Cited by
- ELK4 induced upregulation of HOMER3 promotes the proliferation and metastasis in glioma via Wnt/β-catenin/EMT signaling pathway.
Wang F, Zhou H, Tian Y, Wang X, Huang Y, Tu Y, Li L, Zhen H. Wang F, et al. Biol Direct. 2025 Apr 9;20(1):48. doi: 10.1186/s13062-025-00643-w. Biol Direct. 2025. PMID: 40205485 Free PMC article. - Binge Ethanol Drinking Produces Sexually Divergent and Distinct Changes in Nucleus Accumbens Signaling Cascades and Pathways in Adult C57BL/6J Mice.
Finn DA, Hashimoto JG, Cozzoli DK, Helms ML, Nipper MA, Kaufman MN, Wiren KM, Guizzetti M. Finn DA, et al. Front Genet. 2018 Sep 10;9:325. doi: 10.3389/fgene.2018.00325. eCollection 2018. Front Genet. 2018. PMID: 30250478 Free PMC article. - A role for kalirin in the response of rat medium spiny neurons to cocaine.
Ma XM, Huang JP, Xin X, Yan Y, Mains RE, Eipper BA. Ma XM, et al. Mol Pharmacol. 2012 Oct;82(4):738-45. doi: 10.1124/mol.112.080044. Epub 2012 Jul 24. Mol Pharmacol. 2012. PMID: 22828798 Free PMC article. - The Bermuda Triangle of cocaine-induced neuroadaptations.
Wolf ME. Wolf ME. Trends Neurosci. 2010 Sep;33(9):391-8. doi: 10.1016/j.tins.2010.06.003. Epub 2010 Jul 23. Trends Neurosci. 2010. PMID: 20655604 Free PMC article. Review. - New insights on neurobiological mechanisms underlying alcohol addiction.
Cui C, Noronha A, Morikawa H, Alvarez VA, Stuber GD, Szumlinski KK, Kash TL, Roberto M, Wilcox MV. Cui C, et al. Neuropharmacology. 2013 Apr;67:223-32. doi: 10.1016/j.neuropharm.2012.09.022. Epub 2012 Nov 13. Neuropharmacology. 2013. PMID: 23159531 Free PMC article. Review.
References
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. - PubMed
- Kalivas PW, Volkow N, Seamans J. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–50. - PubMed
- Dom G, Sabbe B, Hulstijn W, van den Brink W. Substance use disorders and the orbitofrontal cortex: systematic review of behavioural decision-making and neuroimaging studies. Br J Psychiatry. 2005;187:209–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AA-016650/AA/NIAAA NIH HHS/United States
- R01 AA016650/AA/NIAAA NIH HHS/United States
- AA-0135017/AA/NIAAA NIH HHS/United States
- AA-015351/AA/NIAAA NIH HHS/United States
- U01 AA016650/AA/NIAAA NIH HHS/United States
- R21 AA015351/AA/NIAAA NIH HHS/United States