mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG - PubMed (original) (raw)
mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG
Hamid Reza Soleimanpour-Lichaei et al. Mol Cell. 2007.
Abstract
Human mitochondria contain their own genome, encoding 13 polypeptides that are synthesized within the organelle. The molecular processes that govern and facilitate this mitochondrial translation remain unclear. Many key factors have yet to be characterized-for example, those required for translation termination. All other systems have two classes of release factors that either promote codon-specific hydrolysis of peptidyl-tRNA (class I) or lack specificity but stimulate the dissociation of class I factors from the ribosome (class II). One human mitochondrial protein has been previously identified in silico as a putative member of the class I release factors. Although we could not confirm the function of this factor, we report the identification of a different mitochondrial protein, mtRF1a, that is capable in vitro and in vivo of terminating translation at UAA/UAG codons. Further, mtRF1a depletion in HeLa cells led to compromised growth in galactose and increased production of reactive oxygen species.
Figures
Figure 1
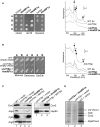
Human mtRF1 Is a Mitochondrial Protein with No Detectable Release Factor Activity (A) Human mtRF1 is targeted to mitochondria. HeLa cells were transiently transfected for 24 hr with a construct expressing 290 N-terminal residues of mtRF1 fused to GFP. Cells were also stained to visualize mitochondria (mitotracker CMH2X-Ros) and nuclei (DAPI). Fluorescence images were captured and mitochondrial colocalization of the fusion protein confirmed by superimposition of the green and red signals in a linescan of the image (line visible in upper panel). An image typical of the three independent transfections is shown. (B) Purified mtRF1 does not induce translation termination in vitro. Human mtRF1 lacking the N-terminal 49 residues was purified and used to assess translation termination from 5 pmol ribosomes programmed with the synthetic codon (400 pmol) indicated. Release factor activity was measured by the hydrolysis of f[3H]met from its cognate tRNAMet as detailed in the Supplemental Experimental Procedures. Nonlimiting amounts of both E. coli RF1 (50 pmol) and UAA triplet (400 pmol) were used in tandem as a positive control for the assay. Standard errors were calculated from a minimum of eight repeats. (C) Human mtRF1 fails to restore respiratory competence in S. cerevisiae Δmrf1. Diploids producing human mtRF1 or containing vector alone were generated from the Δmrf1 strain crossed to the wild-type CW252/A. Following sporulation, asci were dissected and resultant haploid sister spores (A–D) assessed for presence of the plasmid (−uracil), resistance to geneticin G418 showing deletion of MRF1Sc, and growth on a nonfermentable carbon source (glycerol). (D) Human mtRF1 fails to restore respiratory competence in S. pombe Δmrf1. The new fission yeast strain devoid of endogenous mitochondrial release factor NB329 (Experimental Procedures) and its isogenic wild-type strain NB205-6A were transformed with either empty vector or one producing human mtRF1. Transformants were patched on uracil-free minimal medium selecting for the plasmid, replicated onto complete galactose or glycerol plates, and incubated at 28°C.
Figure 2
Human mtRF1a Is a Mitochondrial Release Factor that Recognizes UAA and UAG Codons (A) Human mtRF1a is targeted to mitochondria. HeLa cells were transiently transfected (24 hr) with a construct expressing 357 N-terminal residues of mtRF1a fused to GFP. Cells were also stained to visualize mitochondria (mitotracker CMH2X-Ros) and nuclei (DAPI). Fluorescence images were captured and mitochondrial colocalization of the fusion protein confirmed by superimposition of the green (fusion protein) and red (mitochondrial) signals as shown in the far right panel. The image shown is representative of three independent repeats. (B) Human mtRF1a is imported into mitochondria and matured. Full-length 35S-radiolabeled mtRF1a was in vitro synthesized (lane 1) and incubated with rat liver mitochondria. Under import conditions, two products are visible, the full-length preprotein and the mature protein (lane 2). Components (FCCP, Proteinase K) were added as indicated. (C) Human mtRF1a is found in the mitochondria of all cell lines and tissues analyzed. Human Hep G2, HEK293, and HeLa cell (C60 μg) or mitochondrial (M20 μg) lysates along with human skeletal muscle mitochondria (SkM–M18 μg) were prepared and subjected to western blot analysis with anti-mtRF1a antibodies. A single protein of approximately 40 kDa was visible in all cell/tissue types tested and found exclusively in the mitochondrial fraction (not in postmitochondrial supernatants, data not shown). No crossreactivity was noted with purified mtRF1 (data not shown). (D) Human mtRF1a has release factor activity with UAA and UAG, but not UGA codons. Release assays were performed with 50 pmol mtRF1a and 5 pmol ribosomes, as detailed in the Experimental Procedures. Ribosomes were programmed independently with five triplet codons (400 pmol), four of which are termination codons in human mitochondria (UAA/G, AGA/G), and UGA that encodes tryptophan in mitochondria but acts as a termination codon in the cytosol. No codon controls were also performed. Standard errors were calculated from a minimum of three or a maximum of 11 repeats.
Figure 3
Human mtRF1a Can Suppress the Respiratory Deficiency in Δmrf1 Budding and Fission Yeast (A) Human mtRF1a restores respiratory growth of S. cerevisiae lacking a mitochondrial release factor. The heterozygous Δ_mrf1_ diploid strain (Figure 1C) was transfected with empty vector or encoding human mtRF1a prior to sporulation and tetrad analysis as described. The right panel shows low-temperature spectra of cells grown on glucose (Δ_mrf1_) or glycerol (WT and Δ_mrf1_ + HmtRF1a) medium. (B) Human mtRF1a can function in fission yeast mitochondria. The Δmrf1Sp strain NB329 or the isogenic wild-type was transformed with either control vector or one producing mtRF1a. Transformants patched on uracil-free medium were replicated onto galactose or glycerol/ethanol medium and incubated at 28°C. Spectral analysis from Δ_mrf1_ cells (glucose), WT, and Δmrf1cells expressing MTRF1a (galactose) is shown in the right panel. (C) Human mtRF1a partially restores the level of mitochondrial gene products in yeast. (Left panel) Western analysis of S. cerevisiae respiratory complex subunits Cox2, Atp4, Cyt b from wild-type + empty vector (lane 1), Δmrf1Sc + empty vector (lane 2), Δmrf1Sc + human mtRF1a (lane 3), and Δmrf1Sc + Mrf1Sc (lane 4). The right panel shows total cell extracts of the S. pombe Δ_mrf1_ strain transformed with either the empty vector (lane 1), or plasmids producing the human mtRF1a (lane 2) or Mrf1Sp (lane 3) proteins, and analyzed by western blotting with antibodies recognizing the S. pombe Cox2 or human Hsp60 protein as a loading control. (D) Mitochondrial translation is restored in the presence of human mtRF1a. This was measured with 35S-methionine/cysteine incorporation as detailed in the Experimental Procedures. Strains were as follows: lane 1, Δ_mrf1Sc_ NB345 + vector encoding human mtRF1a; lane 2, Δ_mrf1Sc_ NB345 + empty vector; lane 3, WT NB346 + empty vector.
Figure 4
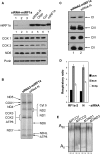
Depletion of Human mtRF1a Does Not Grossly Affect Human Mitochondrial Protein Synthesis (A) Steady-state levels of mtDNA-encoded proteins are unaffected by depletion of mtRF1a. HeLa cells were exposed to siRNA molecules targeting three different regions of the mtRF1a-encoding mRNA sequence (lanes 1–3) or to an untargeted siRNA (siRNA-N, lane 4) for 6 days, and cell lysates (30 μg) were prepared. Control lysates were made from oligofectamine alone (lane 5) and untreated cells (Con, lane 6). Western blots were performed with antibodies against mitochondrial translation products (COX1 cytochrome c oxidase subunit 1; COX2, cytochrome c oxidase subunit II; ND6 NADH CoQ oxidoreductase subunit 6). Porin was used as a nuclear encoded control to confirm equal loading. Purified mtRF1a (5 ng) is loaded in lane 7. The anti-mtRF1a experiment was overexposed to show the level of depletion. The blot accurately reflects three independent experiments. (B) De novo synthesis of mitochondrial proteins is unaffected by depletion of mtRF1a. Cytosolic protein synthesis of HeLa cells grown in the presence of siRNA targeted to MTRF1a (siRNA2-mtRF1a) or untargeted (siRNA-N) for 4 days was inhibited by the addition of emetine, allowing the incorporation of 35S-labeled met/cys directly into the 13 de novo-synthesized mitochondrial proteins. Equal amounts of total cell protein (20 μg) were separated through a 15% SDS PAG and exposed to a PhosphorImager as described. Polypeptides were designated based on their mobilities as described in Chomyn (1996). (C) OXPHOS complexes are present at normal steady-state levels in cells depleted of mtRF1a. HeLa cells were exposed to mtRF1a targeted (lane 1) or untargeted (lane 2) siRNA for 6 days prior to digitonin permeabilisation and preparation for BN-PAGE as described. Following separation, complexes were identified using antisera as detailed. Complex I, anti-39kDa antibody; complex II, anti-SDH 70 kDa antibody; complex III, anti-core 2 antibody; complex IV, anti-COX1 antibody. (D) Respiratory coupling is unaffected in cells depleted of human mtRF1a. HeLa cells (1–2 × 106) grown in targeted (siRNA2-mtRF1a) and untargeted (siRNA-N) siRNA for 3 days were subjected to high-resolution respirometry as detailed. Standard errors were calculated from three measures of respiratory control and capacity. UCR compares the fully uncoupled and resting respiratory rates to give an indication of the respiratory reserve and RCR compares the fully uncoupled respiratory rates to the oligomycin inhibited state 4 respiratory rate, while RCRp combines these ratios to indicate the level of phosphorylation-related respiratory capacity. (E) The poly(A) tail of mitochondrial mRNA is unaffected by depletion of mtRF1a. RNA was isolated from HeLa cells depleted of mtRF1a (lanes 1, siRNA1; lanes 2, siRNA2; lanes 3, siRNA3) and untargeted control siRNA treated (lanes 4) before the poly(A) tail length was assessed for several mt-mRNAs as previously described (Temperley et al., 2003). Poly(A) profiles are shown for transcripts MTCOI, MTCOII, MTND1, and MTND3.
Figure 5
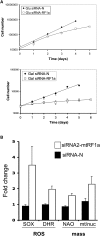
Depletion of mtRF1a Causes a Growth Defect, Increased Mitochondrial ROS Production, and Mitochondrial Mass (A) Cell growth is compromised by depletion of mtRF1a. Multiple aliquots of HeLa cells were exposed to targeted (siRNA2-mtRF1a) and nontargeted (siRNA-N) interfering RNAs for up to 6 days in glucose (promoting glycolysis) or galactose (forcing oxidative respiration) media with cell counts performed each day. Cell numbers are presented in a semi-log plot. The numbers represent a mean ± SEM from four independent wells. (B) Superoxide levels and mitochondrial mass are elevated in cells depleted of mtRF1a. HeLa cells were exposed to targeted (white) and nontargeted (black) siRNA. Superoxide (SOX) was measured with MitoSOX probes and peroxide (DHR) with dihydrorhodamine 123. Levels were measured at day 3 and compared to levels in untreated control cells. The fold increase is shown as a mean ± SEM from at least three independent experiments (p < 0.01 comparing nontargeted to targeted values for mitoSOX and DHR). Mitochondrial mass per cell was measured with the cardiolipin selective dye, NAO. Changes in mtDNA content relative to the nuclear DNA were calculated by qPCR of the MTND1 and 18S rDNA genes and were normalized to untreated cells.
Similar articles
- Structure based hypothesis of a mitochondrial ribosome rescue mechanism.
Huynen MA, Duarte I, Chrzanowska-Lightowlers ZM, Nabuurs SB. Huynen MA, et al. Biol Direct. 2012 May 8;7:14. doi: 10.1186/1745-6150-7-14. Biol Direct. 2012. PMID: 22569235 Free PMC article. - Terminating human mitochondrial protein synthesis: a shift in our thinking.
Lightowlers RN, Chrzanowska-Lightowlers ZM. Lightowlers RN, et al. RNA Biol. 2010 May-Jun;7(3):282-6. doi: 10.4161/rna.7.3.12023. Epub 2010 May 8. RNA Biol. 2010. PMID: 20458175 Review. - HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG.
Nozaki Y, Matsunaga N, Ishizawa T, Ueda T, Takeuchi N. Nozaki Y, et al. Genes Cells. 2008 May;13(5):429-38. doi: 10.1111/j.1365-2443.2008.01181.x. Genes Cells. 2008. PMID: 18429816 - Human mtRF1 terminates COX1 translation and its ablation induces mitochondrial ribosome-associated quality control.
Nadler F, Lavdovskaia E, Krempler A, Cruz-Zaragoza LD, Dennerlein S, Richter-Dennerlein R. Nadler F, et al. Nat Commun. 2022 Oct 27;13(1):6406. doi: 10.1038/s41467-022-34088-w. Nat Commun. 2022. PMID: 36302763 Free PMC article. - Decoding the Enigma: Translation Termination in Human Mitochondria.
Krüger A, Kovalchuk D, Shiriaev D, Rorbach J. Krüger A, et al. Hum Mol Genet. 2024 May 22;33(R1):R42-R46. doi: 10.1093/hmg/ddae032. Hum Mol Genet. 2024. PMID: 38779770 Free PMC article. Review.
Cited by
- Structure based hypothesis of a mitochondrial ribosome rescue mechanism.
Huynen MA, Duarte I, Chrzanowska-Lightowlers ZM, Nabuurs SB. Huynen MA, et al. Biol Direct. 2012 May 8;7:14. doi: 10.1186/1745-6150-7-14. Biol Direct. 2012. PMID: 22569235 Free PMC article. - C6orf203 is an RNA-binding protein involved in mitochondrial protein synthesis.
Gopalakrishna S, Pearce SF, Dinan AM, Schober FA, Cipullo M, Spåhr H, Khawaja A, Maffezzini C, Freyer C, Wredenberg A, Atanassov I, Firth AE, Rorbach J. Gopalakrishna S, et al. Nucleic Acids Res. 2019 Sep 26;47(17):9386-9399. doi: 10.1093/nar/gkz684. Nucleic Acids Res. 2019. PMID: 31396629 Free PMC article. - Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies.
Smits P, Smeitink J, van den Heuvel L. Smits P, et al. J Biomed Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. Epub 2010 Apr 13. J Biomed Biotechnol. 2010. PMID: 20396601 Free PMC article. Review. - A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome.
Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Richter R, et al. EMBO J. 2010 Mar 17;29(6):1116-25. doi: 10.1038/emboj.2010.14. Epub 2010 Feb 25. EMBO J. 2010. PMID: 20186120 Free PMC article. - Host Genetic Risk Factors for Chlamydia trachomatis-Related Infertility in Women.
Zheng X, Zhong W, O'Connell CM, Liu Y, Haggerty CL, Geisler WM, Anyalechi GE, Kirkcaldy RD, Wiesenfeld HC, Hillier SL, Steinkampf MP, Hammond KR, Fine J, Li Y, Darville T. Zheng X, et al. J Infect Dis. 2021 Aug 16;224(12 Suppl 2):S64-S71. doi: 10.1093/infdis/jiab149. J Infect Dis. 2021. PMID: 34396400 Free PMC article.
References
- Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
- Andersson S.G., Zomorodipour A., Andersson J.O., Sicheritz-Pontén T., Alsmark U.C., Podowski R.M., Näslund A.K., Eriksson A.S., Winkler H.H., Kurland C.G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
- Antonicka H., Sasarman F., Kennaway N.G., Shoubridge E.A. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum. Mol. Genet. 2006;15:1835–1846. - PubMed
- Askarian-Amiri M.E., Pel H.J., Guevremont D., McCaughan K.K., Poole E.S., Sumpter V.G., Tate W.P. Functional characterization of yeast mitochondrial release factor 1. J. Biol. Chem. 2000;275:17241–17248. - PubMed
- Barrell B.G., Bankier A.T., Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases