Modification of cellular autophagy protein LC3 by poliovirus - PubMed (original) (raw)
Modification of cellular autophagy protein LC3 by poliovirus
Matthew P Taylor et al. J Virol. 2007 Nov.
Abstract
Poliovirus infection remodels intracellular membranes, creating a large number of membranous vesicles on which viral RNA replication occurs. Poliovirus-induced vesicles display hallmarks of cellular autophagosomes, including delimiting double membranes surrounding the cytosolic lumen, acquisition of the endosomal marker LAMP-1, and recruitment of the 18-kDa host protein LC3. Autophagy results in the covalent lipidation of LC3, conferring the property of membrane association to this previously microtubule-associated protein and providing a biochemical marker for the induction of autophagy. Here, we report that a similar modification of LC3 occurs both during poliovirus infection and following expression of a single viral protein, a stable precursor termed 2BC. Therefore, one of the early steps in cellular autophagy, LC3 modification, can be genetically separated from the induction of double-membraned vesicles that contain the modified LC3, which requires both viral proteins 2BC and 3A. The existence of viral inducers that promote a distinct aspect of the formation of autophagosome-like membranes both facilitates the dissection of this cellular process and supports the hypothesis that this branch of the innate immune response is directly subverted by poliovirus.
Figures
FIG. 1.
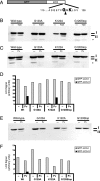
Modification of LC3 protein during poliovirus infection. (A) Schematic representation of LC3 processing as it occurs during autophagy activation. LC3-I is the unmodified, cleaved form of LC3, while LC3-II is the lipid-conjugated species. (B and C) Human 293T cells were either infected with poliovirus at a multiplicity of infection of 50 PFU per cell and harvested at 0, 2, 4, or 6 h after infection, mock infected (-), treated with 10 μM tamoxifen, or treated with control solvent (dimethyl sulfoxide [DMSO]) for 48 h prior to harvesting. (B) Cell extracts were subjected to PAGE through a 13.5% gel, and separated proteins were immunoblotted using a rabbit polyclonal anti-LC3 antibody and subsequently detected with goat anti-rabbit alkaline phosphatase and ECF reagent (Amersham Biosciences). The same immunoblots were reprobed with a mouse monoclonal anticalnexin antibody. (C) The amount of signal for both endogenous LC3-I and endogenous LC3-II from panel B is presented graphically. (D) Cells were transfected with a plasmid that expressed GFP-LC3 for 48 h before infection. Cell extracts were subjected to PAGE in a 10% polyacrylamide-SDS gel, and separated proteins were immunoblotted for LC3 and calnexin as for panel B. The asterisk signifies an unidentified cross-reacting band. (E) The amount of signal for GFP-LC3-I and GFP-LC3-II from panel D is presented graphically. The gels are representative of multiple experiments.
FIG. 2.
Effect of mutations in GFP-LC3 on modification during autophagy induction and poliovirus infection. (A) Schematic depiction of the GFP-LC3 fusion protein sequence. The solid arrowhead indicates the site of cleavage by cellular protease Atg4. Gly120 is the known site of phosphatidylethanolamine addition during autophagy induction. (B) DNA plasmids that encode wild-type GFP-LC3 and variant GFP-LC3 sequences containing G120A, G120Stop, and K122A mutations were transfected into 293T cells. Following transfection, cells were fed with either normal medium (-) or medium that contained 10 μM tamoxifen for 48 h. Cell extracts were displayed on a 10% polyacrylamide-SDS gel and immunoblotted using an anti-LC3 antibody. The asterisk signifies an unidentified cross-reacting band. (C) Wild-type and G120A, K122A, and G120Stop mutant GFP-LC3-encoding plasmids were transfected into 293T cells. After 48 h, cells were infected with poliovirus (50 PFU/cell) and harvested after 6 h. GFP-LC3 protein was detected as for panel B. Gels are representative of multiple experiments. (D) Quantitation of data in panel C is shown. The anti-LC3 immunoblot in panel C was reprobed with an antibody to GFP (E) to test whether the relative immunogenicity of GFP-LC3-I and GFP-LC3-II changed with different antibodies. (F) Relative amounts of GFP-LC3-I and GFP-LC3-II are quantified.
FIG. 3.
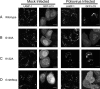
Effect of mutations in GFP-LC3 on its colocalization with LAMP-1 during poliovirus infection. The intracellular distributions of cellular LAMP-1, visualized by indirect immunofluorescence, and GFP-LC3, visualized by its intrinsic fluorescence, were studied following poliovirus infection. Localization patterns of LAMP-1 and wild-type GFP-LC3 (A), G120A GFP-LC3 (B), K122A GFP-LC3 (C), or G120Stop GFP-LC3 (D) in the absence (mock infected) and presence of poliovirus infection are shown. HeLa cells were transfected with GFP-LC3-encoding plasmids prior to infection with poliovirus as in Fig. 2. Cells were fixed 4 h postinfection and stained for LAMP-1 protein with subsequent detection with goat anti-mouse antibody labeled with rhodamine. Bars, 10 μm.
FIG. 4.
Saponin extraction can be used to extract unmodified LC3 species. Human 293T and monkey COS-7 cells were mock infected or infected with poliovirus (50 PFU/cell) for 6 h. Cells were harvested, collected by centrifugation, and subjected to saponin extraction with 0, 0.5, 1, and 2% saponin-containing buffer, as described in Materials and Methods. Proteins were displayed by electrophoresis in 10% acrylamide-SDS gels and visualized by blotting with anti-LC3 or anticalnexin antibodies. The relative amounts of GFP-LC3-I and GFP-LC3-II proteins remaining after saponin extraction were quantified by normalizing the signal in the anti-LC3 immunoblot with the amount of calnexin visualized in the anticalnexin immunoblot.
FIG. 5.
Effect of expression of individual viral proteins on the accumulation of saponin-resistant GFP-LC3-I and GFP-LC3-II proteins. COS-7 cells were transfected with identical total amounts of DNA plasmids, in mixtures that contained GFP-LC3-expressing plasmids and increasing amounts of DNA that expressed 2BC (A), 2B (B), 2C (C), or 3A (D). The balance of the DNA samples comprised the VSV-G-expressing vector into which the poliovirus sequences were cloned. Cells were incubated for 48 h, collected by centrifugation, and subjected to extraction with 0.5% saponin. Saponin-resistant proteins were displayed by electrophoresis in 10% polyacrylamide gels, except as indicated, and probed with three different antibodies for each panel: anti-LC3, to detect GFP-LC3-I and GFP-LC3-II; anti-VSV-G, to normalize for transfection efficiency and toxicity; and anti-2B, anti-2C, or anti-3A, to monitor the expression of the viral protein of interest. (A) Effect of increasing amounts of poliovirus 2BC protein expression. (B) Effect of 2B protein expression. The gel used to resolve the 2B protein was 13.5% acrylamide. (C) Effect of 2C protein expression. (D) Effect of 3A protein expression, compared to the amount of GFP-LC3-I and GFP-LC3-II that was saponin resistant following poliovirus infection as for Fig. 1. The lower portion of each panel shows the amount of saponin-resistant GFP-LC3-I and GFP-LC3-II detected divided by the amount of VSV-G observed and normalized to the control transfection in which no poliovirus protein was expressed. Some toxicity was observed for the greatest amounts of DNA transfected.
FIG. 6.
Comparison of saponin-depleted and nondepleted cell extracts during poliovirus infection and 2BC expression. (A) COS-7 cells were transfected with GFP-LC3 expression plasmid and then 48 h later infected with poliovirus (50 PFU/cell). Cells were then harvested at 0, 2, 4, and 6 h postinfection. (B) COS-7 cells were transfected with a plasmid that expressed GFP-LC3 and increasing proportions of 2BC-expressing plasmids relative to a control vector, followed by harvesting. Recovered cell samples were split in half and either subjected to extraction first with 0.5% saponin and then with NP-40-containing lysis buffer or directly treated with NP-40-containing lysis buffer. Cell extracts were separated by SDS-PAGE on a 10% polyacrylamide gel, transferred to PVDF, and blotted with anti-LC3 and then anti-VSV-G antibodies.
FIG. 7.
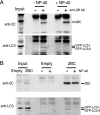
Immunoprecipitation of 2BC from cellular homogenates. COS-7 cells were transfected with both GFP-LC3 and 2BC expression plasmids (A and B) or with empty vector (B). Forty-eight hours later, cells were harvested and lysed by filter homogenization. A low-speed-centrifugation supernatant was collected and used as input for immunoprecipitation. (A) Half of the samples were supplemented with NP-40 and immunoprecipitated with antibody as indicated. (B) One sample of each transfection was supplemented with NP-40 prior to antibody addition to all samples. Immunoprecipitates were separated on an 8% Laemmli gel, transferred to PVDF, and blotted with anti-2C and then anti-LC3 antibodies. Exposure of the LC3 Western blot in panel A is lighter for the input to facilitate visualization of bands.
Similar articles
- Potential subversion of autophagosomal pathway by picornaviruses.
Taylor MP, Kirkegaard K. Taylor MP, et al. Autophagy. 2008 Apr;4(3):286-9. doi: 10.4161/auto.5377. Epub 2007 Dec 5. Autophagy. 2008. PMID: 18094610 Review. - Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm.
Kirkegaard K, Jackson WT. Kirkegaard K, et al. Autophagy. 2005 Oct-Dec;1(3):182-4. doi: 10.4161/auto.1.3.2065. Epub 2005 Oct 30. Autophagy. 2005. PMID: 16874042 - The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein.
Crotty S, Saleh MC, Gitlin L, Beske O, Andino R. Crotty S, et al. J Virol. 2004 Apr;78(7):3378-86. doi: 10.1128/jvi.78.7.3378-3386.2004. J Virol. 2004. PMID: 15016860 Free PMC article. - Subversion of cellular autophagosomal machinery by RNA viruses.
Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Jackson WT, et al. PLoS Biol. 2005 May;3(5):e156. doi: 10.1371/journal.pbio.0030156. Epub 2005 Apr 26. PLoS Biol. 2005. PMID: 15884975 Free PMC article. - LC3 conjugation system in mammalian autophagy.
Tanida I, Ueno T, Kominami E. Tanida I, et al. Int J Biochem Cell Biol. 2004 Dec;36(12):2503-18. doi: 10.1016/j.biocel.2004.05.009. Int J Biochem Cell Biol. 2004. PMID: 15325588 Free PMC article. Review.
Cited by
- Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication.
Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. Romero-Brey I, et al. PLoS Pathog. 2012;8(12):e1003056. doi: 10.1371/journal.ppat.1003056. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236278 Free PMC article. - Crohn disease: a current perspective on genetics, autophagy and immunity.
Stappenbeck TS, Rioux JD, Mizoguchi A, Saitoh T, Huett A, Darfeuille-Michaud A, Wileman T, Mizushima N, Carding S, Akira S, Parkes M, Xavier RJ. Stappenbeck TS, et al. Autophagy. 2011 Apr;7(4):355-74. doi: 10.4161/auto.7.2.13074. Epub 2011 Apr 1. Autophagy. 2011. PMID: 20729636 Free PMC article. Review. - Manipulation of autophagy by (+) RNA viruses.
Wong HH, Sanyal S. Wong HH, et al. Semin Cell Dev Biol. 2020 May;101:3-11. doi: 10.1016/j.semcdb.2019.07.013. Epub 2019 Aug 8. Semin Cell Dev Biol. 2020. PMID: 31382014 Free PMC article. Review. - Dengue virus-induced autophagy regulates lipid metabolism.
Heaton NS, Randall G. Heaton NS, et al. Cell Host Microbe. 2010 Nov 18;8(5):422-32. doi: 10.1016/j.chom.2010.10.006. Cell Host Microbe. 2010. PMID: 21075353 Free PMC article. - Eating the enemy within: autophagy in infectious diseases.
Orvedahl A, Levine B. Orvedahl A, et al. Cell Death Differ. 2009 Jan;16(1):57-69. doi: 10.1038/cdd.2008.130. Epub 2008 Sep 5. Cell Death Differ. 2009. PMID: 18772897 Free PMC article. Review.
References
- Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. - PubMed
- Bilir, A., M. A. Altinoz, M. Erkan, V. Ozmen, and A. Aydiner. 2001. Autophagy and nuclear changes in FM3A breast tumor cells after epirubicin, medroxyprogesterone and tamoxifen treatment in vitro. Pathobiology 69:120-126. - PubMed
- Bursch, W., A. Ellinger, H. Kienzl, L. Torok, S. Pandey, M. Sikorska, R. Walker, and R. S. Hermann. 1996. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis 17:1595-1607. - PubMed
- Bursch, W., K. Hochegger, L. Torok, B. Marian, A. Ellinger, and R. S. Hermann. 2000. Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J. Cell Sci. 113:1189-1198. - PubMed
- Chang, C. P., M. C. Yang, H. S. Liu, Y. S. Lin, and H. Y. Lei. 2007. Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology 45:286-296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous