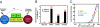A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist - PubMed (original) (raw)
A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
W David Tolbert et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Hepatocyte growth factor (HGF) activates the Met receptor tyrosine kinase by binding and promoting receptor dimerization. Here we describe a mechanistic basis for designing Met antagonists based on NK1, a natural variant of HGF containing the N-terminal and the first kringle domain. Through detailed biochemical and structural analyses, we demonstrate that both mouse and human NK1 induce Met dimerization via a conserved NK1 dimer interface. Mutations designed to alter the NK1 dimer interface abolish its ability to promote Met dimerization but retain full Met-binding activity. Importantly, these NK1 mutants act as Met antagonists by inhibiting HGF-mediated cell scattering, proliferation, branching, and invasion. The ability to separate the Met-binding activity of NK1 from its Met dimerization activity thus provides a rational basis for designing Met antagonists. This strategy of antagonist design may be applicable for other growth factor receptors by selectively abolishing the receptor activation ability but not the receptor binding of the growth factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Binding of the human and mouse NK1 to Met. (A) A schematic representation of the domain arrangement of HGF and Met. (B) The purified proteins used in this study are depicted. Proteins shown are the mouse and human NK1 (lanes 1 and 2), the human NK1 with R134G mutation (lane 3), biotinylated human NK1 (lane 4), and the Met sema domain (lane 5). The asterisk in lane 2 indicates the truncated N domain of human NK1. The dashed line indicates that lanes 4 and 5 were run in a separate gel. (C) A diagram of AlphaScreen assay for detecting NK1-Met interactions. (D) Binding of biotinylated NK1 to Met in the absence or presence of heparin as measured by AlphaScreen assay is depicted. (E) Binding affinity (IC50 values) of various NK1s to Met as determined by the inhibition of the binding of the biotinylated NK1 to Met with dose competition of unlabeled NK1s is shown.
Fig. 2.
Met dimerization induced by NK1 binding. (A) A diagram of AlphaScreen assay for detecting Met dimerization promoted by NK1 binding. (B) Met dimerization induced by 1 μM NK1 in the presence and absence of 10 μM heparin as measured by AlphaScreen assays. (C) Dose curves of various NK1 to induce Met dimerization in the presence of 10 μM heparin.
Fig. 3.
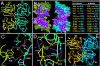
Crystal structures of the mouse and human NK1. (A) An overall view of the head-to-tail dimer of the mouse NK1. The letters (K, D, Y, and N) indicate the position of residues K85, D123, Y124, and N127, respectively. Only yellow monomer is labeled. (B) The NK1 dimer interface and the intermolecular interactions. The interface within 6.0 Å of the other monomer is shown as a purple surface and inner residues are gold, green, and red if they are within 4.0, 5.0, and 6.0 Å, respectively, of the other NK1. The list of interactions includes non-H-bond packing and H-bond interactions within 4.0 Å between the two NK1 monomers. Double-headed arrows indicate reciprocal interactions between two monomers and single-headed arrows indicate interactions from one monomer to the other. Underlining indicates that the residues' protein backbone atoms are involved in dimer interactions. (C) Interactions mediated by Y124 and K85 (yellow monomer) with V140, D202, I203, and P204 (cyan monomer). Hydrogen bonds are indicated by dashed lines. Water-mediated interactions are indicated by W1 and W2. (D) Interactions mediated by K122, D123, and N127 (yellow monomer) with K122, D123, and N127 (cyan monomer). Hydrogen bonds are indicated by dashed lines. (E) Comparison of the mouse and human NK1 (superposition of the Cα atoms). The two mouse monomers are in yellow and cyan, and the human monomers are in gold and blue. Arrows indicate the heparin binding sites identified in previous studies (23).
Fig. 4.
Effects of the NK1 dimerization mutants. (A) The ability of various NK1 mutants to induce Met dimerization in the presence and absence of 10 μM heparin. The D123A and N127A single mutants behave similarly to the K85A mutant (data not shown). (B) The ability of NK1 mutants (1 μM) to inhibit NK1-mediated Met dimerization.
Fig. 5.
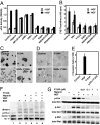
Effects of NK1 dimerization mutants in cell-based assays. (A) Effects of NK1 mutants (1 μM) on uPA assays in the presence and absence of HGF. (B) Effects of NK1 mutants (1 μM) on proliferation of MDCK cells in the presence and absence of HGF. (C) Effects of NK1 (1 μM) on MDCK cell-scattering assays in the presence and absence of HGF. (D) Effects of NK1 mutants (1 μM) on branching morphogenesis of the prostate cancer cell line DU145. Arrows indicate the cell branching induced by HGF. (E) Effects of NK1 mutants (1 μM) on invasion of the glioblastoma cell line DBTRG. (F) Inhibition of Met dimerization on the DU145 cell surface by the Y124A mutant as detected by Western blot analysis of cross-linking proteins. The positions of Met monomer (p145) and dimer as well as the unprocessed Met (p175) and the nonspecific band are indicated. (G) Inhibition of Akt and Erk phosphorylation by the Y124A mutant. All assays were performed with 60–100 ng/ml HGF when indicated and 2 μM heparin except for F and G, where heparin used is 1.0 mg/ml.
Similar articles
- Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1.
Niemann HH. Niemann HH. Biochim Biophys Acta. 2013 Oct;1834(10):2195-204. doi: 10.1016/j.bbapap.2012.10.012. Epub 2012 Oct 31. Biochim Biophys Acta. 2013. PMID: 23123275 Review. - Crystal structures of NK1-heparin complexes reveal the basis for NK1 activity and enable engineering of potent agonists of the MET receptor.
Lietha D, Chirgadze DY, Mulloy B, Blundell TL, Gherardi E. Lietha D, et al. EMBO J. 2001 Oct 15;20(20):5543-55. doi: 10.1093/emboj/20.20.5543. EMBO J. 2001. PMID: 11597998 Free PMC article. - Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2.
Schwall RH, Chang LY, Godowski PJ, Kahn DW, Hillan KJ, Bauer KD, Zioncheck TF. Schwall RH, et al. J Cell Biol. 1996 May;133(3):709-18. doi: 10.1083/jcb.133.3.709. J Cell Biol. 1996. PMID: 8636243 Free PMC article. - Structural basis for agonism and antagonism of hepatocyte growth factor.
Tolbert WD, Daugherty-Holtrop J, Gherardi E, Vande Woude G, Xu HE. Tolbert WD, et al. Proc Natl Acad Sci U S A. 2010 Jul 27;107(30):13264-9. doi: 10.1073/pnas.1005183107. Epub 2010 Jul 12. Proc Natl Acad Sci U S A. 2010. PMID: 20624990 Free PMC article. - Structural insights into Met receptor activation.
Niemann HH. Niemann HH. Eur J Cell Biol. 2011 Nov;90(11):972-81. doi: 10.1016/j.ejcb.2010.11.014. Epub 2011 Jan 15. Eur J Cell Biol. 2011. PMID: 21242015 Review.
Cited by
- Plexin structures are coming: opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions.
Hota PK, Buck M. Hota PK, et al. Cell Mol Life Sci. 2012 Nov;69(22):3765-805. doi: 10.1007/s00018-012-1019-0. Epub 2012 Jun 29. Cell Mol Life Sci. 2012. PMID: 22744749 Free PMC article. Review. - Allosteric peptide activators of pro-hepatocyte growth factor stimulate Met signaling.
Landgraf KE, Santell L, Billeci KL, Quan C, Young JC, Maun HR, Kirchhofer D, Lazarus RA. Landgraf KE, et al. J Biol Chem. 2010 Dec 17;285(51):40362-72. doi: 10.1074/jbc.M110.179721. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937841 Free PMC article. - Flexibility and small pockets at protein-protein interfaces: New insights into druggability.
Jubb H, Blundell TL, Ascher DB. Jubb H, et al. Prog Biophys Mol Biol. 2015 Oct;119(1):2-9. doi: 10.1016/j.pbiomolbio.2015.01.009. Epub 2015 Feb 7. Prog Biophys Mol Biol. 2015. PMID: 25662442 Free PMC article. Review. - Engineering hepatocyte growth factor fragments with high stability and activity as Met receptor agonists and antagonists.
Jones DS 2nd, Tsai PC, Cochran JR. Jones DS 2nd, et al. Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):13035-40. doi: 10.1073/pnas.1102561108. Epub 2011 Jul 25. Proc Natl Acad Sci U S A. 2011. PMID: 21788476 Free PMC article. - Single-molecule photobleaching reveals increased MET receptor dimerization upon ligand binding in intact cells.
Dietz MS, Haße D, Ferraris DM, Göhler A, Niemann HH, Heilemann M. Dietz MS, et al. BMC Biophys. 2013 Jun 3;6(1):6. doi: 10.1186/2046-1682-6-6. BMC Biophys. 2013. PMID: 23731667 Free PMC article.
References
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. - PubMed
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Nature. 1995;376:768–771. - PubMed
- Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Science. 1991;251:802–804. - PubMed
- Cioce V, Csaky KG, Chan AM, Bottaro DP, Taylor WG, Jensen R, Aaronson SA, Rubin JS. J Biol Chem. 1996;271:13110–13115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK066202/DK/NIDDK NIH HHS/United States
- DK071662/DK/NIDDK NIH HHS/United States
- G9704528/MRC_/Medical Research Council/United Kingdom
- DK066202/DK/NIDDK NIH HHS/United States
- R01 DK071662/DK/NIDDK NIH HHS/United States
- G0501019/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous