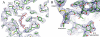Crystal structure of a thermally stable rhodopsin mutant - PubMed (original) (raw)
Crystal structure of a thermally stable rhodopsin mutant
Jörg Standfuss et al. J Mol Biol. 2007.
Abstract
We determined the structure of the rhodopsin mutant N2C/D282C expressed in mammalian cells; the first structure of a recombinantly produced G protein-coupled receptor (GPCR). The mutant was designed to form a disulfide bond between the N terminus and loop E3, which allows handling of opsin in detergent solution and increases thermal stability of rhodopsin by 10 deg.C. It allowed us to crystallize a fully deglycosylated rhodopsin (N2C/N15D/D282C). N15 mutations are normally misfolding and cause retinitis pigmentosa in humans. Microcrystallographic techniques and a 5 microm X-ray beam were used to collect data along a single needle measuring 5 microm x 5 microm x 90 microm. The disulfide introduces only minor changes but fixes the N-terminal cap over the beta-sheet lid covering the ligand-binding site, a likely explanation for the increased stability. This work allows structural investigation of rhodopsin mutants and shows the problems encountered during structure determination of GPCRs and other mammalian membrane proteins.
Figures
Figure 1. Western blot of rhodopsin mutants and three-dimensional crystals of recombinant rhodopsins
A: The proteins were expressed and purified from transfected COS cells. Primary antibody used in the blot was 1D4. Lane 1: The N2C/D282C mutant has lost the carbohydrate chain on residue 2 but retains that attached to residue 15. Lane 2: The fully deglycosylated mutant N2C/N15D/D282C shows a single homogeneous band. Mutations in position 15 are known to cause autosomal dominant retinitis pigmentosa in humans. Photos from crystals of recombinant rhodopsins were taken under infrared illumination (>850nm). B: Crystals of recombinant wild type rhodopsin. C: Crystals of thermally stabilized rhodopsin N2C/D282C. D: Crystals of stabilized fully deglycosylated rhodopsin N2C/N15D/D282C.
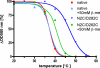
Figure 2. Thermal stability of N2C/D282C rhodopsin in the detergent C8E4 used for crystallization
The N2C/D282C mutant was purified by 1D4 immunoaffinity chromatography. Rhodopsin solubilized from retinas (native) was purified by Con A affinity chromatography. In both cases a Sephadex G50 column was used to exchange the detergent from 0.02% (w/v) DDM to 0.2% (w/v) C8E4 immediately before the experiment. The protein solutions were incubated for 30 minutes at various temperatures before an absorption spectrum has been recorded. The amount of bound retinal was determined by the absorption at 500 nm, corrected by the protein absorption at 280 nm. To investigate the effect of reduced disulfide bonds the same experiment was performed in the presence of 50 mM β-mercaptoethanol (β-me). Using the program Prism sigmoidal curves were fitted to the individual data points. The T50 values obtained from these curves (wt: 37.3°C., wt+β-me: 37.5°C., N2C/D282C: 47.2°C., N2C/D282C+β-me: 41.3°C.) show a ∼10°C. increase in stability of the mutant which is mostly lost in the presence of β-mercaptoethanol.
Figure 3. Data collection on a recombinant microcrystal
A: Frozen microcrystal used for data collection. The arrow represents the axis along which the data was collected on 15 distinct positions. B: Diffraction pattern obtained after 15 s exposure and 1° rotation. 1: The first diffraction pattern collected at a new position on the needle. 2: The eighth consecutive frame collected at the same position. A strong reduction in diffraction intensity caused by radiation damage is observed. 3: Fully restored diffraction intensity is obtained after the beam had been translated along the needle axis. C: Diffraction intensity represented by the number of strong spots (NSTRONG, XDS) on each frame of the 13 indexed wedges. At each minimum in the number of strong spots the beam was translated to a new position on the needle axis. The positions of diffraction patterns shown in B are marked.
Figure 4. Electron density maps of N2C/D282C rhodopsin
A: Retinal Omit electron density map (0.75 sigma) obtained after molecular replacement and density modification. The retinal (red) was not included but appears clearly in the electron density. B: Disulfide bond and N15 glycosylation. Sigma-A weighted 2Fo-Fc Electron density contoured at sigma 0.9. The disulfide bond between the mutated residues C2 and C282 is well resolved, as is the first residue of the N-glycosylation linked to N15. No density is visible adjacent to the mutated residue 2, which would have been glycosylated in wild-type rhodopsin.
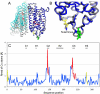
Figure 5. Comparison of recombinant and native rhodopsin
A: Superposition emphasizing overall similarity of the individual monomers and their different dimer packing along helix 5. Blue: Monomer A, Cyan: Monomer B, Grey: native rhodopsin monomers A and B obtained from 1GZM coordinates, Yellow: Designed disulfide bond between C2 and C282, Red:retinal, Green: N15 linked glycosylation B: Enlargement of the extracellular region with the introduced disulfide bond. C: Coordinate differences between monomers A of both structures plotted as the root mean square deviation (rmsd) of Cα atoms against sequence position (blue). Poorly ordered regions with a B-factor higher than 100 are marked red. The designed disulfide includ ing two amino acids on each side is labeled yellow. The largest differences are found in loops C2 and C3 and are due to the inherent flexibility and poor order of these loops. The end of helix 5 before the C3 loop is shifted about 2 Å due to the different dimer packing. The disulfide is introducing small but significant changes in the backbone of the E3 loop and the N-terminus.
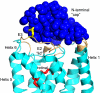
Figure 6. The N-terminal Cap domain
Contacts between the N-terminal cap (blue) and the rest of the receptor (cyan). Atoms of the receptor with a minimum distance of 5 Å to residues 1-33 of the N-terminus are colored beige. The covalently bound retinal (red) is separated from the N-terminal cap by the lid formed from the E2 loop. The designed disulfide between C2 and C282 connecting the N-terminal cap with loop E3 is shown as yellow sticks. The molecule is viewed with the extracellular site facing up.
Similar articles
- An opsin mutant with increased thermal stability.
Xie G, Gross AK, Oprian DD. Xie G, et al. Biochemistry. 2003 Feb 25;42(7):1995-2001. doi: 10.1021/bi020611z. Biochemistry. 2003. PMID: 12590586 - Retinitis pigmentosa mutants provide insight into the role of the N-terminal cap in rhodopsin folding, structure, and function.
Opefi CA, South K, Reynolds CA, Smith SO, Reeves PJ. Opefi CA, et al. J Biol Chem. 2013 Nov 22;288(47):33912-33926. doi: 10.1074/jbc.M113.483032. Epub 2013 Oct 8. J Biol Chem. 2013. PMID: 24106275 Free PMC article. - Structure and function in rhodopsin: packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain are coupled.
Hwa J, Garriga P, Liu X, Khorana HG. Hwa J, et al. Proc Natl Acad Sci U S A. 1997 Sep 30;94(20):10571-6. doi: 10.1073/pnas.94.20.10571. Proc Natl Acad Sci U S A. 1997. PMID: 9380676 Free PMC article. - Relevance of rhodopsin studies for GPCR activation.
Deupi X. Deupi X. Biochim Biophys Acta. 2014 May;1837(5):674-82. doi: 10.1016/j.bbabio.2013.09.002. Epub 2013 Sep 13. Biochim Biophys Acta. 2014. PMID: 24041646 Review. - G protein-coupled receptor drug discovery: implications from the crystal structure of rhodopsin.
Ballesteros J, Palczewski K. Ballesteros J, et al. Curr Opin Drug Discov Devel. 2001 Sep;4(5):561-74. Curr Opin Drug Discov Devel. 2001. PMID: 12825452 Free PMC article. Review.
Cited by
- Low aqueous solubility of 11-cis-retinal limits the rate of pigment formation and dark adaptation in salamander rods.
Frederiksen R, Boyer NP, Nickle B, Chakrabarti KS, Koutalos Y, Crouch RK, Oprian D, Cornwall MC. Frederiksen R, et al. J Gen Physiol. 2012 Jun;139(6):493-505. doi: 10.1085/jgp.201110685. J Gen Physiol. 2012. PMID: 22641642 Free PMC article. - The minimum crystal size needed for a complete diffraction data set.
Holton JM, Frankel KA. Holton JM, et al. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66(Pt 4):393-408. doi: 10.1107/S0907444910007262. Epub 2010 Mar 24. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20382993 Free PMC article. - Optimisation of Neuraminidase Expression for Use in Drug Discovery by Using HEK293-6E Cells.
Campbell AC, Tanner JJ, Krause KL. Campbell AC, et al. Viruses. 2021 Sep 22;13(10):1893. doi: 10.3390/v13101893. Viruses. 2021. PMID: 34696326 Free PMC article. - GPCR: G protein complexes--the fundamental signaling assembly.
Jastrzebska B. Jastrzebska B. Amino Acids. 2013 Dec;45(6):1303-14. doi: 10.1007/s00726-013-1593-y. Epub 2013 Sep 20. Amino Acids. 2013. PMID: 24052187 Free PMC article. Review. - Molecular basis of cannabinoid CB1 receptor coupling to the G protein heterotrimer Gαiβγ: identification of key CB1 contacts with the C-terminal helix α5 of Gαi.
Shim JY, Ahn KH, Kendall DA. Shim JY, et al. J Biol Chem. 2013 Nov 8;288(45):32449-32465. doi: 10.1074/jbc.M113.489153. Epub 2013 Oct 3. J Biol Chem. 2013. PMID: 24092756 Free PMC article.
References
- Drews J, Ryser S. The role of innovation in drug development. Nat Biotechnol. 1997;15:1318–9. - PubMed
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. - PubMed
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–83. - PubMed
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY007965-16/EY/NEI NIH HHS/United States
- R01 EY007965/EY/NEI NIH HHS/United States
- R01 EY007965-17/EY/NEI NIH HHS/United States
- EY007965/EY/NEI NIH HHS/United States
- MC_U105178937/MRC_/Medical Research Council/United Kingdom
- R01 EY007965-18/EY/NEI NIH HHS/United States
- R01 EY007965-19/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous