Drosophila Myosin II, Zipper, is essential for ommatidial rotation - PubMed (original) (raw)
Drosophila Myosin II, Zipper, is essential for ommatidial rotation
Ryan W Fiehler et al. Dev Biol. 2007.
Abstract
The adult Drosophila retina is a highly polarized epithelium derived from a precursor tissue that is initially symmetric across its dorsoventral axis. Specialized 90 degrees rotational movements of subsets of cells, the ommatidial precursors, establish mirror symmetry in the retinal epithelium. Myosin II, or Zipper (Zip), a motor protein, regulates the rate at which ommatidia rotate: in zip mutants, the rate of rotation is significantly slowed. Zip is concentrated in the cells that we show to be at the likely interface between rotating and non-rotating cells: the boundary between differentiated and undifferentiated cells. Zip is also robust in newly added ommatidial cells, consistent with our model that the machinery that drives rotation should shift to newly recruited cells as they are added to the growing ommatidium. Finally, cell death genes and canonical Wnt signaling pathway members genetically modify the zip phenotype.
Figures
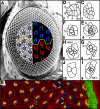
Figure 1. The Drosophila eye is a polarized epithelium
(A) Scanning electron micrograph of a Drosophila compound eye. A tangential demi-section reveals the underlying cellular morphology of the eye. The rhabdomeres, or light-sensitive organelles of the photoreceptors, form chiral trapezoids that are shown schematically in blue (dorsally-oriented trapezoids) and red (ventrally-oriented trapezoids). The trapezoids are oriented perpendicular to the equator, the line of mirror symmetry that divides the eye into dorsal and ventral halves (yellow line). Arrow highlights ommatidia separated by 4°. (B) Apical surface of a third instar eye imaginal disc immunostained with α-Arm and pseudocolored to highlight the morphogenetic furrow (green), arcs (orange), photoreceptors R8, R2, R5, R1, R7 and R6 (orange), and R3 and R4 (blue). The direction of rotation is indicated by blue arrows. The interface between rotating cells could reside between undifferentiated, interommatidial cells (white and green asterisks) or between rotating photoreceptors (red dots) and stationary undifferentiated cells (yellow asterisks). (C-J) Video lucida drawings of a maturational time line of ommatidial precursors. See text for details. Numbers in lower left hand corners indicate row number. M1, M2: mystery cells; 1-8: photoreceptor cells; a.c. anterior cone cell; p.c. posterior cone cell; pol.c.: polar cone cell; eq.c.: equatorial cone cell. Anterior to the right.
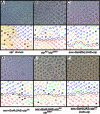
Figure 2. zip exhibits an orientation phenotype
Clones of the null allele zip1 (A) and hypomorphic transheterozygous flies (zipEbr/zip02957) (B) exhibit rotation errors. In (A), shaded area denotes regions of phenotypically mutant tissue, presumably a consequence of loss of zip mutant cells since no _w_- photoreceptors can be identified. Misexpression of zip (C) and zipDN (D) under the sev promoter results in defects in ommatidial rotation. To demonstrate the efficacy of the sev>zipDN transgene, zip dosage was decreased (E) or increased (F) in a sev>zipDN background. Decreasing zip dosage using zip02957 enhances the sev>zipDN phenotype (E) whereas increasing zip dosage using a second copy of the UAS>zip transgene suppresses the sev>zipDN phenotype. M, missing photoreceptors; E, extra photoreceptors; blue trapezoids: dorsal ommatidia; red trapezoids: ventral ommatidia; green trapezoids: under-rotated ommatidia; orange trapezoids: over-rotated ommatidia.
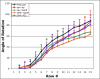
Figure 3. zip regulates the rate of ommatidial rotation
Wild-type rotation begins at row 4 and is effectively complete by row 15 (black). Decreasing zip function using the sev and GMR promoters to drive zipDN slows the rate of rotation (sev>zipDN, green, and GMR>zipDN, orange). Misexpressing wild-type zip in a subset of cells also disrupts the rate of rotation (sev>zip, red). Misexpressing P35 under the GMR promoter delays rotation in a pattern similar to sev>zip (purple). Wild-type is statistically different from: sev>zip in rows 6-8 and 11-12 (t-test, p<.001); sev>zipDN in rows 6 and 9-15 (t-test, p<.001); GMR>zipDN in rows 5-15 (t-test, p<.001); GMR>P35 in rows 5-14 (t-test, p<.001).
Figure 4. The interface for rotation resides between the photoreceptors/cone cells and undifferentiated cells
(A) and (B): Two models illustrating potential interfaces between rotating cells. In (A) like-colored cells are linked together and move as a group so the interface is at the purple/orange border. In (B), undifferentiated cells do not rotate with differentiated cells. The interface between rotating and stationary cells is between the colored and uncolored cells. (C) Average angle of orientation by developmental stage. Each “stick diagram” corresponds to the ommatidial form that lies directly above or below the diagram. In the diagrams, the center line indicates the average angle, red shading highlights standard deviation, and outermost lines indicate minimum (lower line) and maximum (higher line) degree of rotation seen for each developmental stage indicated. (D) and (E): Schematic representation of relative likelihood of anterior and posterior cone cells being recruited from the BUdR-labeled pool of cells. Green-colored gradient represents probability of a cell being labeled with BUdR. In both panels, two ommatidial precursors are shown, one anterior to and the second posterior to, the BUdR-labeled band. Darker cells are more likely to be labeled since they lie closer to this band. Numbers identify cells that will become anterior and posterior cone cells. In (D), cells are recruited prior to rotation (as modeled in Fig. 4B), so cells numbered 1 through 4, which have equivalent levels of shading, share equal probabilities of being labeled. When examined as pupal eyes, equal numbers of labeled anterior and posterior cells are predicted. In (E), cells are recruited after rotation begins (as modeled in Fig. 4C). Consequently, two cells are recruited from the pool that is more likely to be labeled (6,7, dark green) whereas the other two cells are recruited from the pool that is less likely to be labeled (5,8, pale green). When examined as pupal eyes, the number of labeled anterior and posterior cells is predicted to be disproportionate. See text for details. Anterior is to the right for all panels.
Figure 5. Positions and fates adopted by cells in second mitotic wave
Each of the three schematics represents an independent eye. Black cells denote cells in S phase at the time a pulse of BUdR was administered. Whereas the anterior and posterior cone cells rotate with the photoreceptors, the future pigment cells - the undifferentiated, interommatidial cells of the third instar - retain their positions (i.e. they do not rotate with the photoreceptors). The interface between rotating and non-rotating cells therefore lies between the photoreceptors/cone cells and the interommatidial cells. See text for details and inset for cell types. 1-6: photoreceptor cells; B: bristle; 1°, 2° and 3°: primary, secondary and tertiary pigment cells, respectively; Po: polar cone cell; Eq: equatorial cone cell; A: anterior cone cell; P: posterior cone cell. Arrows denote the position of the equator. Anterior is to the right.
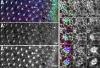
Figure 6. Zip localizes strongly to differentiating cells
(A-F) Third larval instar eye disc stained with α-Arm (green), α-Zip (red), and phalloidin (blue). Co-localization of Zip (A’-F’) and actin (A”-F”) is evident in ommatidia before (arrowhead, A-A”) and during (arrow, A-A”) rotation. Close-ups of ommatidia (B-F) show dynamic co-localization of both actin and Zip. Accumulations are seen at the interface between ommatidial and undifferentiated cells (arrowheads C’, C”, E’, E”) and in the cytoplasm of rotating photoreceptors (arrows, E’ and E”). Anterior is to the right.
Figure 7. sev>zipDN interacts with canonical Wnt signaling and cell death genes
sev>zipDN is dominantly suppressed by dsh1 (A), but no interaction is seen with the tissue polarity gene stbm6cn (B). However sev>zipDN is enhanced by fzKD4A (C) and dsh3 (D) (compare to figure 2D). An allele of arm (arm8) (E) specific for canonical Wnt signaling also enhances sev>zipDN. GMR>P35 (F) produces an ommatidial orientation phenotype, and a deficiency that removes grim, rpr, hid (Df (H99)) (G) and hid05014 (H) suppress sev>zipDN. M, missing photoreceptors; E, extra photoreceptors; blue trapezoids: dorsal ommatidia; red trapezoids: ventral ommatidia; green trapezoids: under-rotated ommatidia; orange trapezoids: over-rotated ommatidia.
Figure 8. The interommatidial lattice is disrupted when Zip function is compromised
(A) Wild-type mid-pupal eye stained with α-Arm. The interommatidial lattice in wild-type eyes is virtually error-free. 1°, 2°, 3°: primary, secondary, and tertiary pigment cells, respectively; cc: cone cells. Bristles cells appear as white circles in these preparations. zipEbr/zip02957 (B) and sev>zipDN (C) mid-pupal eyes stained with α -Arm. Multiple patterning defects arise when Zip function is decreased. See text for details.
Similar articles
- Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation.
Fiehler RW, Wolff T. Fiehler RW, et al. Dev Biol. 2008 Jan 15;313(2):533-44. doi: 10.1016/j.ydbio.2007.10.034. Epub 2007 Oct 30. Dev Biol. 2008. PMID: 18068152 Free PMC article. - EGF signaling and ommatidial rotation in the Drosophila eye.
Strutt H, Strutt D. Strutt H, et al. Curr Biol. 2003 Aug 19;13(16):1451-7. doi: 10.1016/s0960-9822(03)00545-1. Curr Biol. 2003. PMID: 12932331 - Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila.
Wolff T, Rubin GM. Wolff T, et al. Development. 1998 Mar;125(6):1149-59. doi: 10.1242/dev.125.6.1149. Development. 1998. PMID: 9463361 - Planar cell polarity signaling in the Drosophila eye.
Jenny A. Jenny A. Curr Top Dev Biol. 2010;93:189-227. doi: 10.1016/B978-0-12-385044-7.00007-2. Curr Top Dev Biol. 2010. PMID: 20959167 Free PMC article. Review. - NOTCH and the patterning of ommatidial founder cells in the developing Drosophila eye.
Baker NE. Baker NE. Results Probl Cell Differ. 2002;37:35-58. doi: 10.1007/978-3-540-45398-7_4. Results Probl Cell Differ. 2002. PMID: 25707068 Review. No abstract available.
Cited by
- Dynamics of cell polarity in tissue morphogenesis: a comparative view from Drosophila and Ciona.
Veeman MT, McDonald JA. Veeman MT, et al. F1000Res. 2016 Jun 2;5:F1000 Faculty Rev-1084. doi: 10.12688/f1000research.8011.1. eCollection 2016. F1000Res. 2016. PMID: 27303647 Free PMC article. Review. - Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila.
O'Keefe DD, Gonzalez-Niño E, Burnett M, Dylla L, Lambeth SM, Licon E, Amesoli C, Edgar BA, Curtiss J. O'Keefe DD, et al. Dev Biol. 2009 Sep 1;333(1):143-60. doi: 10.1016/j.ydbio.2009.06.032. Epub 2009 Jul 1. Dev Biol. 2009. PMID: 19576205 Free PMC article. - Nemo kinase phosphorylates β-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-β-catenin.
Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, Mlodzik M. Mirkovic I, et al. Nat Struct Mol Biol. 2011 Jun;18(6):665-72. doi: 10.1038/nsmb.2049. Epub 2011 May 8. Nat Struct Mol Biol. 2011. PMID: 21552260 Free PMC article. - Integrins are required for synchronous ommatidial rotation in the Drosophila eye linking planar cell polarity signalling to the extracellular matrix.
Thuveson M, Gaengel K, Collu GM, Chin ML, Singh J, Mlodzik M. Thuveson M, et al. Open Biol. 2019 Aug 30;9(8):190148. doi: 10.1098/rsob.190148. Epub 2019 Aug 14. Open Biol. 2019. PMID: 31409231 Free PMC article. - Myosin II regulates extension, growth and patterning in the mammalian cochlear duct.
Yamamoto N, Okano T, Ma X, Adelstein RS, Kelley MW. Yamamoto N, et al. Development. 2009 Jun;136(12):1977-86. doi: 10.1242/dev.030718. Epub 2009 May 13. Development. 2009. PMID: 19439495 Free PMC article.
References
- Ahmed Y, et al. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell. 1998;93:1171–82. - PubMed
- Ahmed Y, et al. Drosophila Apc1 and Apc2 regulate Wingless transduction throughout development. Development. 2002;129:1751–62. - PubMed
- Baker NE, et al. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6:1290–301. - PubMed
- Barros CS, et al. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–40. - PubMed
- Baumann O. Spatial pattern of nonmuscle myosin-II distribution during the development of the Drosophila compound eye and implications for retinal morphogenesis. Dev Biol. 2004;269:519–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases