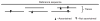Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans - PubMed (original) (raw)
. 2007 Oct;39(10):1251-5.
doi: 10.1038/ng2116. Epub 2007 Sep 9.
Affiliations
- PMID: 17828266
- PMCID: PMC3586588
- DOI: 10.1038/ng2116
Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans
Alon Keinan et al. Nat Genet. 2007 Oct.
Abstract
Large data sets on human genetic variation have been collected recently, but their usefulness for learning about history and natural selection has been limited by biases in the ways polymorphisms were chosen. We report large subsets of SNPs from the International HapMap Project that allow us to overcome these biases and to provide accurate measurement of a quantity of crucial importance for understanding genetic variation: the allele frequency spectrum. Our analysis shows that East Asian and northern European ancestors shared the same population bottleneck expanding out of Africa but that both also experienced more recent genetic drift, which was greater in East Asians.
Figures
Figure 1
Discovery of SNPs by comparing two sequencing reads from an individual of known ancestry. SNPs useable for analysis are identified as sites where one read matches the reference sequence and the other does not (an arrowhead of either type indicates a mismatch compared with the reference sequence; see Methods). The leftmost SNP is not ascertained because there is only one read, and the rightmost SNP is not ascertained because both reads share the same allele.
Figure 2
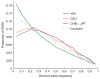
Derived allele frequency spectra in each population. The derived allele frequency spectrum (the proportion of SNPs of each possible derived allele frequency) is shown for each of the HapMap populations, after discovery of SNPs in two reads of the same ancestry. The YRI spectrum is based on SNPs ascertained in both the Cor17109 and the Cor17119 libraries; the CEU spectrum is based on the Cor7340 and the HuAA libraries; and the CHB+JPT spectrum is based on the Cor11321 and the HuFF libraries. SNPs ascertained in individuals of the same ancestry are pooled together (Supplementary Fig. 1 online), as are allele frequency data from the two East Asian populations, CHB and JPT (Supplementary Fig. 2 online). Also shown is the expected derived allele frequency spectrum for a population of constant size throughout history and the same ascertainment scheme. Although all spectra are biased by discovery in two chromosomes, they are comparable because the bias is identical for all spectra (we account for this bias in our analyses; see Methods).
Figure 3
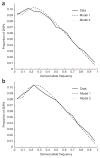
Modeling provides an excellent fit to the observed allele frequency spectra. (a,b) CEU (a) and CHB+JPT (b) data compared with that predicted by the models. Model 1 allows for one bottleneck in the history of each population, and model 2 allows for two bottlenecks (Supplementary Methods). For presentation (not actual analysis), the spectra are divided into 15 bins. Model 2 provides a better fit to the data, with mean squared error (averaged over bins) reduced to 62% of the model 1 value in CEU and 47% of the model 1 value in CHB+JPT.
Similar articles
- GWAS-identified schizophrenia risk SNPs at TSPAN18 are highly diverged between Europeans and East Asians.
Liu J, Li M, Su B. Liu J, et al. Am J Med Genet B Neuropsychiatr Genet. 2016 Dec;171(8):1032-1040. doi: 10.1002/ajmg.b.32471. Epub 2016 Jun 17. Am J Med Genet B Neuropsychiatr Genet. 2016. PMID: 27312590 - Genetic evidence for the convergent evolution of light skin in Europeans and East Asians.
Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. Norton HL, et al. Mol Biol Evol. 2007 Mar;24(3):710-22. doi: 10.1093/molbev/msl203. Epub 2006 Dec 20. Mol Biol Evol. 2007. PMID: 17182896 - Empirical distributions of F(ST) from large-scale human polymorphism data.
Elhaik E. Elhaik E. PLoS One. 2012;7(11):e49837. doi: 10.1371/journal.pone.0049837. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185452 Free PMC article. - The genetic architecture of normal variation in human pigmentation: an evolutionary perspective and model.
McEvoy B, Beleza S, Shriver MD. McEvoy B, et al. Hum Mol Genet. 2006 Oct 15;15 Spec No 2:R176-81. doi: 10.1093/hmg/ddl217. Hum Mol Genet. 2006. PMID: 16987881 Review. - Associations between ERAP1 polymorphisms and susceptibility to ankylosing spondylitis: a meta-analysis.
Lee YH, Song GG. Lee YH, et al. Clin Rheumatol. 2016 Aug;35(8):2009-2015. doi: 10.1007/s10067-016-3287-9. Epub 2016 Apr 25. Clin Rheumatol. 2016. PMID: 27108589 Review.
Cited by
- Demographic inference using spectral methods on SNP data, with an analysis of the human out-of-Africa expansion.
Lukic S, Hey J. Lukic S, et al. Genetics. 2012 Oct;192(2):619-39. doi: 10.1534/genetics.112.141846. Epub 2012 Aug 3. Genetics. 2012. PMID: 22865734 Free PMC article. - Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations.
Kim BY, Lohmueller KE. Kim BY, et al. Am J Hum Genet. 2015 Mar 5;96(3):454-61. doi: 10.1016/j.ajhg.2014.12.029. Epub 2015 Feb 12. Am J Hum Genet. 2015. PMID: 25683122 Free PMC article. - The genomic landscape of Neanderthal ancestry in present-day humans.
Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. Sankararaman S, et al. Nature. 2014 Mar 20;507(7492):354-7. doi: 10.1038/nature12961. Epub 2014 Jan 29. Nature. 2014. PMID: 24476815 Free PMC article. - Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution.
Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Fumagalli M, et al. PLoS Genet. 2011 Nov;7(11):e1002355. doi: 10.1371/journal.pgen.1002355. Epub 2011 Nov 3. PLoS Genet. 2011. PMID: 22072984 Free PMC article. - The deleterious mutation load is insensitive to recent population history.
Simons YB, Turchin MC, Pritchard JK, Sella G. Simons YB, et al. Nat Genet. 2014 Mar;46(3):220-4. doi: 10.1038/ng.2896. Epub 2014 Feb 9. Nat Genet. 2014. PMID: 24509481 Free PMC article.
References
- The International HapMap Consortium. The Phase II HapMap. in the press.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials