USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier - PubMed (original) (raw)
USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier
Suming Huang et al. Mol Cell Biol. 2007 Nov.
Abstract
The insulator element at the 5' end of the chicken beta-globin locus acts as a barrier, protecting transgenes against silencing effects of adjacent heterochromatin. We showed earlier that the transcription factor USF1 binds within the insulator and that this site is important for generating in adjacent nucleosomes histone modifications associated with active chromatin and, by inference, with barrier function. To understand the mechanism of USF1 action, we have characterized USF1-containing complexes. USF1 interacts directly with the histone H4R3-specific methyltransferase PRMT1. USF1, PRMT1, and the histone acetyltransferases (HATs) PCAF and SRC-1 form a complex with both H4R3 histone methyltransferase and HAT activities. Small interfering RNA downregulation of USF1 results in localized loss of H4R3 methylation, and other histone modifications associated with euchromatin, at the insulator. A dominant negative peptide that interferes with USF1 binding to DNA causes silencing of an insulated reporter construct, indicating abolition of barrier function. These results show that USF1 plays a direct role in maintaining the barrier, supporting a model in which the insulator works as a barrier by maintaining a local environment of active chromatin.
Figures
FIG. 1.
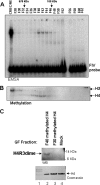
Histone methyltransferases associate with the FIV DNA-binding complex. (A) Fractionation of nuclear extract from chicken erythrocytes (CRBC/NE). Column fractions derived from the Sephacryl S-300 HR column were analyzed. An electrophoretic mobility shift assay with column fractions using 32P-labeled FIV oligonucleotide duplexes shows that USF1 DNA-binding activity is present in two complexes of approximately 1.8 MDa and 180 to 400 kDa by gel filtration. (B) Fluorogram of the same column fractions used in the HMT assay. Each USF1 complex coelutes with different HMT activities with specificity for H3 or H4 separately. (C) Association of histone H4R3 methylation with USF1 activity. Recombinant histone H4 was subjected to methylation with column fractions number 30 and 40, representing two different USF1 DNA-binding complexes. (Top) Western blotting analysis with a dimethyl H4R3-specific antibody. (Bottom) Coomassie stain of recombinant histone H4 and core histone isolated from chicken used in the Western blot assay.
FIG. 2.
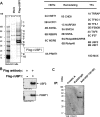
Identification of the USF1-interacting proteins. (A) USF1-interacting polypeptides were purified from 1 × 109 FLAG-HA-tagged USF1-transduced and mock-transduced HeLa cells by using a FLAG antibody-conjugated column. The polypeptides were eluted by the FLAG peptide and were resolved by SDS-PAGE. The proteins were visualized with Coomassie blue staining and were analyzed by mass spectrometry. A partial list of polypeptides identified is listed on the right. (B) The FLAG-purified samples from FLAG-USF1-transduced and mock-transduced cells were analyzed by immunoblotting with antibodies against USF1, USF2, and PRMT1. (C) USF1 interacts with PRMT1 in chicken erythroleukemia 6C2 cells. Nuclear extracts from 6C2 cells were immunoprecipitated with protein A beads, rabbit IgG, and USF1 antibody. The precipitated samples were analyzed by immunoblotting with PRMT1 antibody.
FIG. 3.
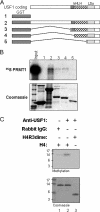
USF1 directly recruits PRMT1 and H4R3 HMT activity. (A) The HLH domain of USF1 interacts with Arg 3-specific methyltransferase PRMT1 directly. Shown is a schematic representation of the GST-USF1 fusion proteins used in the GST pull-down assay. (B) 35S-labeled PRMT1 was incubated with GST and GST-USF1 fusion proteins preadsorbed to glutathione-Sepharose beads. (Top) Bound PRMT1 was eluted and visualized by fluorography following SDS-PAGE. (Bottom) A Coomassie-stained gel shows the relative amounts of fusion proteins used in the assay. (C) USF1 recruits H4R3-specific HMT. Nuclear extracts from HeLa cells were immunoprecipitated with rabbit IgG or USF1 antibody. The precipitates were incubated with unmethylated H4 peptide (lane 1 and 2) or R3 dimethylated H4 peptide (lane 3) in the presence of [3H]AdoMet as cofactor. The proteins were resolved by SDS-PAGE and were visualized by fluorography (top). The loading amounts of H4 used in the assay were demonstrated by Coomassie blue staining (bottom).
FIG. 4.
USF1 forms a complex with PRMT1 and HATs. Nuclear extracts from HeLa cells expressing FLAG-USF1 were fractionated through a Sephacryl S-300 HR column. (A to E) Fractions were collected and were analyzed by immunoblotting with antibodies against USF1 (A), PRMT1 (B), PCAF (C), SRC-1 (D), and p300 (E). (F to K) The peak fractions from 1.8 MDa (I), 400 kDa (II), and <50 kDa (III) were collected. The pooled fractions were immunoprecipitated with FLAG antibody. The precipitates were then analyzed by Western blot analysis using anti-USF1, anti-USF2, anti-PRMT1, anti-PCAF, and anti-SRC-1 antibodies. (L) HeLa nuclear extracts were immunoprecipitated with rabbit IgG or USF1 antibody and then were incubated with chicken core histones in the presence of [3H]acetyl coenzyme A (CoA). Histones were resolved by SDS-PAGE and were visualized by fluorography (top) or Coomassie blue staining (bottom).
FIG. 5.
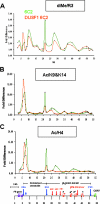
Knockdown of USF1 leads to decreased methylation at H4R3 as well as acetylation of H3 and H4. USF1 expression was reduced 80 to 90% in 6C2 cells based on RNA interference technology (42). The results of ChIP assays with antibodies specific to dimethylated H4R3 (A), acetylated H3K9/K14 (B), and acetylated H4 (C) from wild-type (green) or USF1 knockdown (orange) 6C2 cell line are shown. See Materials and Methods for a description of the measurements and calculations of the difference.
FIG. 6.
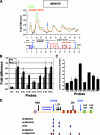
USF1 is required for histone modifications at the 5′HS4 insulator element. (A) Results of chromatin IP with antibodies specific for dimethyl H3K9 from wild-type (green) and USF1 knockdown (orange) 6C2 cell lines across 54 kb of the chicken β-globin locus. (B) Focusing on histone H3K27 trimethylation at the chicken β-globin domain following USF1 knockdown. ChIP of trimethylated H3K27 at endogenous sequences in wild-type (WT; gray bars) or USF1 knockdown (black bars) cells. (C) The relative enrichment of histone H3 K27 trimethylation is shown across the β-globin domain by comparing the USF1 knockdown to wild-type 6C2 cells. (D) Summary of the ChIP data across the chicken β-globin locus, showing the results of the USF1 knockdown. The arrows pointing up and down indicate increases or decreases in the modification, respectively.
FIG. 7.
USF1 is responsible for preventing chromosomal position-effect silencing. (A) Schematic representation of the insulated IL-2R transgene, which was amplified four times (four copies) in the 6C2 genome (line 809). (B) Line 809 was transfected with vector containing AUSF1 cDNA, which lacks the basic DNA-binding region and inhibits both USF1 and USF2 activities. The single clones (B7 and C11) were selected in the presence of 100 μg/ml zeocin (Invitrogen). Gel mobility shift assays of nuclear extracts from the control and AUSF1 clones were carried out using 32P-labeled FIV oligonucleotide duplexes. (C) Results of the FACS analysis for IL-2R expression at 0 and 70 days after removal of hygromycin selection. M1 designates IL-2R-positive cells. On these histograms, cell number is indicated on the y axis and the logarithm of fluorescein isothiocyanate fluorescence intensity is on the x axis. (D) Results of the FACS analysis for IL-2R expression of clone B7 (top) and clone C11 (bottom) at 0 and 15 days after removal of hygromycin selection.
FIG. 8.
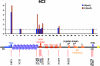
USF1 does not recruit RNA polymerase II to the 5′HS4 insulator at the chicken β-globin locus. A ChIP analysis of Pol II localization at the 54-kb chicken β-globin locus in 6C2 cells is shown. The IPs using anti-Pol II phosphor Ser 2 are indicated by blue bars, and those that used anti-Pol II phosphor Ser 5 are indicated by red bars. RNA polymerase levels were compared to those for a site within the 16-kb condensed chromatin region, which was assumed to be low (see Materials and Methods).
Similar articles
- H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation.
Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S. Li X, et al. Blood. 2010 Mar 11;115(10):2028-37. doi: 10.1182/blood-2009-07-236059. Epub 2010 Jan 12. Blood. 2010. PMID: 20068219 Free PMC article. - Recruitment of histone modifications by USF proteins at a vertebrate barrier element.
West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. West AG, et al. Mol Cell. 2004 Nov 5;16(3):453-63. doi: 10.1016/j.molcel.2004.10.005. Mol Cell. 2004. PMID: 15525517 - Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes.
Li X, Wang S, Li Y, Deng C, Steiner LA, Xiao H, Wu C, Bungert J, Gallagher PG, Felsenfeld G, Qiu Y, Huang S. Li X, et al. Blood. 2011 Aug 4;118(5):1386-94. doi: 10.1182/blood-2010-11-319111. Epub 2011 Jun 8. Blood. 2011. PMID: 21653943 Free PMC article. - Chromatin insulator elements: establishing barriers to set heterochromatin boundaries.
Barkess G, West AG. Barkess G, et al. Epigenomics. 2012 Feb;4(1):67-80. doi: 10.2217/epi.11.112. Epigenomics. 2012. PMID: 22332659 Review. - Interplay between chromatin remodelers and protein arginine methyltransferases.
Pal S, Sif S. Pal S, et al. J Cell Physiol. 2007 Nov;213(2):306-15. doi: 10.1002/jcp.21180. J Cell Physiol. 2007. PMID: 17708529 Review.
Cited by
- Enhancers: the abundance and function of regulatory sequences beyond promoters.
Bulger M, Groudine M. Bulger M, et al. Dev Biol. 2010 Mar 15;339(2):250-7. doi: 10.1016/j.ydbio.2009.11.035. Epub 2009 Dec 16. Dev Biol. 2010. PMID: 20025863 Free PMC article. Review. - H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation.
Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S. Li X, et al. Blood. 2010 Mar 11;115(10):2028-37. doi: 10.1182/blood-2009-07-236059. Epub 2010 Jan 12. Blood. 2010. PMID: 20068219 Free PMC article. - Histone crosstalk directed by H2B ubiquitination is required for chromatin boundary integrity.
Ma MK, Heath C, Hair A, West AG. Ma MK, et al. PLoS Genet. 2011 Jul;7(7):e1002175. doi: 10.1371/journal.pgen.1002175. Epub 2011 Jul 21. PLoS Genet. 2011. PMID: 21811414 Free PMC article. - Role of helix-loop-helix proteins during differentiation of erythroid cells.
Anantharaman A, Lin IJ, Barrow J, Liang SY, Masannat J, Strouboulis J, Huang S, Bungert J. Anantharaman A, et al. Mol Cell Biol. 2011 Apr;31(7):1332-43. doi: 10.1128/MCB.01186-10. Epub 2011 Jan 31. Mol Cell Biol. 2011. PMID: 21282467 Free PMC article. Review. - Locus control region mediated regulation of adult beta-globin gene expression.
Liang S, Moghimi B, Yang TP, Strouboulis J, Bungert J. Liang S, et al. J Cell Biochem. 2008 Sep 1;105(1):9-16. doi: 10.1002/jcb.21820. J Cell Biochem. 2008. PMID: 18500726 Free PMC article.
References
- Abramovich, C., and R. K. Humphries. 2005. HOX regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 12:210-216. - PubMed
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. - PubMed
- Andrews, G. K., D. K. Lee, R. Ravindra, P. Lichtlen, M. Sirito, M. Sawadogo, and W. Schaffner. 2001. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-1 expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 20:1114-1122. - PMC - PubMed
- Barski, A., S. Cuddapah, K. Cui, T.-Y. Ron, D. E. Schones, Z. Wang, G. Wei, L. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. - PubMed
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous