Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia - PubMed (original) (raw)
Case Reports
Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia
Lynn Adaimy et al. Am J Hum Genet. 2007 Oct.
Abstract
Odonto-onycho-dermal dysplasia is a rare autosomal recessive syndrome in which the presenting phenotype is dry hair, severe hypodontia, smooth tongue with marked reduction of fungiform and filiform papillae, onychodysplasia, keratoderma and hyperhidrosis of palms and soles, and hyperkeratosis of the skin. We studied three consanguineous Lebanese Muslim Shiite families that included six individuals affected with odonto-onycho-dermal dysplasia. Using a homozygosity-mapping strategy, we assigned the disease locus to an ~9-cM region at chromosome 2q35-q36.2, located between markers rs16853834 and D2S353, with a maximum multipoint LOD score of 5.7. Screening of candidate genes in this region led us to identify the same c.697G-->T (p.Glu233X) homozygous nonsense mutation in exon 3 of the WNT10A gene in all patients. At the protein level, the mutation is predicted to result in a premature truncated protein of 232 aa instead of 417 aa. This is the first report to our knowledge of a human phenotype resulting from a mutation in WNT10A, and it is the first demonstration of an ectodermal dysplasia caused by an altered WNT signaling pathway, expanding the list of WNT-related diseases.
Figures
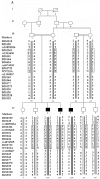
Figure 1.
Pedigrees, haplotypes, and WNT10A mutation status of family 1 (A), family 2 (B), and family 3 (C). Individuals’ numbers are listed below the pedigree symbols. Genotyped markers from the chromosome 2q34-q36.2 region are shown to the left, and individuals’ allele numbers for each marker are given next to the bar. Black bars represent the haplotype segregating with the odonto-onycho-dermal gene. WNT10A is located between markers D2S2722 and rs1109867. The mutation genotypes of WNT10A are shown below haplotypes. + indicates mutation Glu233X; − indicates wild-type allele. A border indicates the common haplotype 1-C-3-3-4-T-1-T between the three families, and the arrow shows the neomutation in patient V:3 of family 3.
Figure 1.
Pedigrees, haplotypes, and WNT10A mutation status of family 1 (A), family 2 (B), and family 3 (C). Individuals’ numbers are listed below the pedigree symbols. Genotyped markers from the chromosome 2q34-q36.2 region are shown to the left, and individuals’ allele numbers for each marker are given next to the bar. Black bars represent the haplotype segregating with the odonto-onycho-dermal gene. WNT10A is located between markers D2S2722 and rs1109867. The mutation genotypes of WNT10A are shown below haplotypes. + indicates mutation Glu233X; − indicates wild-type allele. A border indicates the common haplotype 1-C-3-3-4-T-1-T between the three families, and the arrow shows the neomutation in patient V:3 of family 3.
Figure 1.
Pedigrees, haplotypes, and WNT10A mutation status of family 1 (A), family 2 (B), and family 3 (C). Individuals’ numbers are listed below the pedigree symbols. Genotyped markers from the chromosome 2q34-q36.2 region are shown to the left, and individuals’ allele numbers for each marker are given next to the bar. Black bars represent the haplotype segregating with the odonto-onycho-dermal gene. WNT10A is located between markers D2S2722 and rs1109867. The mutation genotypes of WNT10A are shown below haplotypes. + indicates mutation Glu233X; − indicates wild-type allele. A border indicates the common haplotype 1-C-3-3-4-T-1-T between the three families, and the arrow shows the neomutation in patient V:3 of family 3.
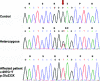
Figure 2.
WNT10A mutation associated with odonto-onycho-dermal dysplasia. Electropherograms of WNT10A exon 3 in a control individual (top), one parent (middle), and an affected patient (bottom). DNA sequence analysis revealed a homozygous G→T substitution at nucleotide 697 (from the translation start site) in the affected patient, causing the Glu233X mutation. The parent is heterozygous with respect to the Glu233X mutation.
Similar articles
- Tricho-odonto-onycho-dermal dysplasia and WNT10A mutations.
Kantaputra P, Kaewgahya M, Jotikasthira D, Kantaputra W. Kantaputra P, et al. Am J Med Genet A. 2014 Apr;164A(4):1041-8. doi: 10.1002/ajmg.a.36388. Epub 2014 Jan 23. Am J Med Genet A. 2014. PMID: 24458874 - Odonto-onycho-dermal dysplasia in a patient homozygous for a WNT10A nonsense mutation and mild manifestations of ectodermal dysplasia in carriers of the mutation.
Krøigård AB, Clemmensen O, Gjørup H, Hertz JM, Bygum A. Krøigård AB, et al. BMC Dermatol. 2016 Mar 10;16:3. doi: 10.1186/s12895-016-0040-7. BMC Dermatol. 2016. PMID: 26964878 Free PMC article. - Further delineation of the odonto-onycho-dermal dysplasia syndrome.
Mégarbané H, Haddad M, Delague V, Renoux J, Boehm N, Mégarbané A. Mégarbané H, et al. Am J Med Genet A. 2004 Aug 30;129A(2):193-7. doi: 10.1002/ajmg.a.30188. Am J Med Genet A. 2004. PMID: 15316967 - The fundamentals of WNT10A.
Benard EL, Hammerschmidt M. Benard EL, et al. Differentiation. 2025 Mar-Apr;142:100838. doi: 10.1016/j.diff.2025.100838. Epub 2025 Jan 30. Differentiation. 2025. PMID: 39904689 Free PMC article. Review. - Ectodermal dysplasia-skin fragility syndrome: Two new cases and review of this desmosomal genodermatosis.
Doolan BJ, Gomaa NS, Fawzy MM, Dogheim NN, Liu L, Mellerio JE, Onoufriadis A, McGrath JA. Doolan BJ, et al. Exp Dermatol. 2020 Jun;29(6):520-530. doi: 10.1111/exd.14096. Epub 2020 May 25. Exp Dermatol. 2020. PMID: 32248567 Review.
Cited by
- The First Report of a Missense Variant in RFX2 Causing Non-Syndromic Tooth Agenesis in a Consanguineous Pakistani Family.
Khan SA, Khan S, Muhammad N, Rehman ZU, Khan MA, Nasir A, Kalsoom UE, Khan AK, Khan H, Wasif N. Khan SA, et al. Front Genet. 2022 Jan 25;12:782653. doi: 10.3389/fgene.2021.782653. eCollection 2021. Front Genet. 2022. PMID: 35145545 Free PMC article. - Wnt and the Wnt signaling pathway in bone development and disease.
Wang Y, Li YP, Paulson C, Shao JZ, Zhang X, Wu M, Chen W. Wang Y, et al. Front Biosci (Landmark Ed). 2014 Jan 1;19(3):379-407. doi: 10.2741/4214. Front Biosci (Landmark Ed). 2014. PMID: 24389191 Free PMC article. Review. - Molecular basis of hypohidrotic ectodermal dysplasia: an update.
Trzeciak WH, Koczorowski R. Trzeciak WH, et al. J Appl Genet. 2016 Feb;57(1):51-61. doi: 10.1007/s13353-015-0307-4. Epub 2015 Aug 21. J Appl Genet. 2016. PMID: 26294279 Free PMC article. Review. - Wnt/beta-catenin signaling: components, mechanisms, and diseases.
MacDonald BT, Tamai K, He X. MacDonald BT, et al. Dev Cell. 2009 Jul;17(1):9-26. doi: 10.1016/j.devcel.2009.06.016. Dev Cell. 2009. PMID: 19619488 Free PMC article. Review. - Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction.
Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R. Zhang Y, et al. Dev Cell. 2009 Jul;17(1):49-61. doi: 10.1016/j.devcel.2009.05.011. Dev Cell. 2009. PMID: 19619491 Free PMC article.
References
Web Resources
- Ensembl, http://www.ensembl.org/index.html
- Genatlas, http://www.genatlas.org
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for EPHA4 [accession number NM_004438], INHA [accession number NM_002191], DNAJB2 [accession number NM_006736], VIL1 [accession number NM_007127], WNT6 [accession number NM_006522], and WNT10A [accession number NM_025216.2])
References
- Itin PH, Fistarol SK (2004) Ectodermal dysplasias. Am J Med Genet C Semin Med Genet 131:45–51 - PubMed
- Freire-Maia N, Pinheiro M (1984) Ectodermal dysplasias: a clinical and genetic study. Alan R. Liss, New York, pp 172–173
- Freire-Maia N, Pinheiro M (1988) Selected conditions with ectodermal dysplasia. Birth Defects Orig Artic Ser 24:109–121 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases