Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements - PubMed (original) (raw)
Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements
Jonathan E Aldridge et al. PLoS One. 2007.
Abstract
In an accompanying paper, we show that the mitochondrial Unfolded Protein Response or mtUPR is initiated by the activation of transcription of chop through an AP-1 element in the chop promoter. Further, we show that the c/ebp beta gene is similarly activated and CHOP and C/EBP beta subsequently hetero-dimerise to activate transcription of mtUPR responsive genes. Here, we report the discovery of six additional mtUPR responsive genes. We found that these genes encoding mitochondrial proteases YME1L1 and MPP beta, import component Tim17A and enzymes NDUFB2, endonuclease G and thioredoxin 2, all contain a CHOP element in their promoters. In contrast, genes encoding mitochondrial proteins Afg3L2, Paraplegin, Lon and SAM 50, which do not have a CHOP element, were not up-regulated. Conversely, genes with CHOP elements encoding cytosolic proteins were not induced by the accumulation of unfolded proteins in mitochondria. These results indicate that mtUPR responsive genes appear to share a requirement for a CHOP element, but that this is not sufficient for the regulation of the mtUPR. A more detailed analysis of promoters of mtUPR responsive genes revealed at least two additional highly conserved, putative regulatory sites either side of the CHOP element, one a motif of 12 bp which lies 14 bp upstream of the CHOP site and another 9 bp element, 2 bp downstream of the CHOP site. Both of these additional elements are conserved in the promoters of 9 of the ten mtUPR responsive genes we have identified so far, the exception being the Cpn60/10 bidirectional promoter. Mutation of each of these elements substantially reduced the mtUPR responsiveness of the promoters suggesting that these elements coordinately regulate mtUPR.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
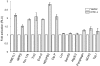
Figure 1. Survey of promoters responsive to mtUPR.
Reporter constructs for mitochondrial proteins YME1L1, MPPβ (
m
itochondrial
p
rocessing
p
eptidase β subunit), Trx2 (mitochondrial
t
hio
r
edo
x
in), Tim17A, End G (
Endo
nuclease
G
), NDUFB2 (subunit of complex I), and ClpP, which have CHOP consensus, and Lon, Sam50, Afg3L2, Paraplegin without CHOP consensus, or NOX3 (
N
ADPH
ox
idases
3
) and Typ I (
Typ
e
I
iodothyronine deiodinase), which are non-mitochondrial proteins but have CHOP consensus sequence in the promoter region, were tested. COS-7 cells were co-transfected with vector or vector containing OTCΔ and promoter-luciferase constructs and were used for luciferase assay 32 h after transfection. Data represent the mean±SEM from experiments performed in triplicate.
Figure 2. Nucleotide sequence alignment of the promoter region of mtUPR responsive genes.
(A): mtUPR Genes, indicating the position in the promoter relative to the transcription start site, are shown on the left hand side of the figure. Asterisks identify identical sequences and bold letters show the highly conserved elements MURE1, CHOP, and MURE2. (B): Consensus sequence of the MURE1 and MURE2 elements, as well as the sequence for CHOP taken from .
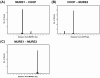
Figure 3. Effect of the mutation on MURE1 and MURE2 elements on mtUPR responsiveness.
A. Effect of MURE1 mutations on yme1l1 promoter activity.; B. Effect of MURE2 mutations on yme1l1 promoter activity; C. Effect of MURE2 mutations on mppβ promoter activity. Luciferase activity is compared for each mutation in cells co-transfected with empty vector or vector- OTCΔ and the promoter reporter constructs and is expressed as the activity relative to that obtained for the wild type promoter (RLU-relative luciferase units). The results represent the mean±SEM for 3 independent experiments. The promoter sequence for wild type and mutants is shown.
Figure 4. Spacing of MURE1, CHOP and MURE2 motifs.
MURE1-CHOP (A), CHOP-MURE2 (B) and MURE1-MURE2 (C). Each figure shows the number of motif pairs with the given separation found in the set of predicted mitochondrial proteins. Separation is defined as the distance from the first base of the first motif to the first base of the second motif.
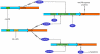
Figure 5. Comparison of regulatory circuits for mtUPR and erUPR.
MtUPR activates transcription of both chop and c/ebpβ genes through an AP-1 site. These 2 transcription factors then bind to a CHOP element in the mtUPR responsive genes, along with transcription factors of unknown identity which bind to the MURE1 and 2 elements. ErUPR also activates transcription of CHOP (but not C/EBP genes) through an ER response element (ERSE). The function of CHOP in erUPR appears to be restricted to the induction of apoptosis through transcriptional activation of BIM.
Similar articles
- The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response.
Horibe T, Hoogenraad NJ. Horibe T, et al. PLoS One. 2007 Sep 12;2(9):e835. doi: 10.1371/journal.pone.0000835. PLoS One. 2007. PMID: 17848986 Free PMC article. - Regulation of aldehyde reductase expression by STAF and CHOP.
Barski OA, Papusha VZ, Kunkel GR, Gabbay KH. Barski OA, et al. Genomics. 2004 Jan;83(1):119-29. doi: 10.1016/s0888-7543(03)00213-1. Genomics. 2004. PMID: 14667815 - Mitochondrial Unfolded Protein Response (mtUPR) and Diseases.
Yang M, Luo S, Chen W, He L, Liu D, Wang X, Xiao L, Sun L. Yang M, et al. Curr Med Chem. 2025;32(9):1674-1684. doi: 10.2174/0929867331666230822095924. Curr Med Chem. 2025. PMID: 37608662 Review. - A New Vision of Mitochondrial Unfolded Protein Response to the Sirtuin Family.
Weng H, Ma Y, Chen L, Cai G, Chen Z, Zhang S, Ye Q. Weng H, et al. Curr Neuropharmacol. 2020;18(7):613-623. doi: 10.2174/1570159X18666200123165002. Curr Neuropharmacol. 2020. PMID: 31976838 Free PMC article. Review.
Cited by
- Molecular mechanism of cytotoxicity induced by Hsp90-targeted Antp-TPR hybrid peptide in glioblastoma cells.
Horibe T, Torisawa A, Kohno M, Kawakami K. Horibe T, et al. Mol Cancer. 2012 Aug 22;11:59. doi: 10.1186/1476-4598-11-59. Mol Cancer. 2012. PMID: 22913813 Free PMC article. - Molecular Determinants of Mitochondrial Shape and Function and Their Role in Glaucoma.
Khalimonchuk O, Becker DF. Khalimonchuk O, et al. Antioxid Redox Signal. 2023 May;38(13-15):896-919. doi: 10.1089/ars.2022.0124. Epub 2023 Jan 5. Antioxid Redox Signal. 2023. PMID: 36301938 Free PMC article. Review. - Altered Mitochondrial Protein Homeostasis and Proteinopathies.
Jishi A, Qi X. Jishi A, et al. Front Mol Neurosci. 2022 Apr 27;15:867935. doi: 10.3389/fnmol.2022.867935. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35571369 Free PMC article. Review. - Are the estrogen receptor and SIRT3 axes of the mitochondrial UPR key regulators of breast cancer sub-type determination according to age?
Jenkins EC, Chattopadhyay M, Germain D. Jenkins EC, et al. Aging Cancer. 2021 Sep;2(3):75-81. doi: 10.1002/aac2.12035. Epub 2021 Aug 6. Aging Cancer. 2021. PMID: 34927079 Free PMC article. - Mitochondrial Quality Control Strategies: Potential Therapeutic Targets for Neurodegenerative Diseases?
Hu D, Liu Z, Qi X. Hu D, et al. Front Neurosci. 2021 Nov 12;15:746873. doi: 10.3389/fnins.2021.746873. eCollection 2021. Front Neurosci. 2021. PMID: 34867159 Free PMC article. Review.
References
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. - PubMed
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. - PubMed
- Schroder M. The unfolded protein response. Mol Biotechnol. 2006;34:279–90. - PubMed
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. - PubMed
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials