A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification - PubMed (original) (raw)
A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification
Dorothee Mueller et al. Blood. 2007.
Abstract
Chimeric proteins joining the histone methyltransferase MLL with various fusion partners trigger distinctive lymphoid and myeloid leukemias. Here, we immunopurified proteins associated with ENL, a protein commonly fused to MLL. Identification of these ENL-associated proteins (EAPs) by mass spectrometry revealed enzymes with a known role in transcriptional elongation (RNA polymerase II C-terminal domain kinase [RNAPolII CTD] positive transcription elongation factor b [pTEFb]), and in chromatin modification (histone-H3 methyltransferase DOT1L) as well as other frequent MLL partners (AF4, AF5q31, and LAF4), and polycomb group members (RING1, CBX8, and BCoR). The composition of EAP was further verified by coimmunoprecipitation, 2-hybrid analysis, pull-down, and colocalization experiments. Purified EAP showed a histone H3 lysine 79-specific methylase activity, displayed a robust RNAPolII CTD kinase function, and counteracted the effect of the pTEFb inhibitor 5,6-dichloro-benzimidazole-riboside. In vivo, an ENL knock-down diminished genome-wide as well as gene-specific H3K79 dimethylation, reduced global run-on elongation, and inhibited transient transcriptional reporter activity. According to structure-function data, DOT1L recruitment was important for transformation by the MLL-ENL fusion derivative. These results suggest a function of ENL in histone modification and transcriptional elongation.
Figures
Figure 1
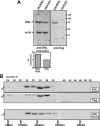
Characterization of an fENL producer cell line. (A) Detection of flagENL expression by immunoblot. Extracts from transduced 293fENL and HEK293 parental cells were tested for endogenous and ectopic ENL expression by probing with anti-ENL and anti-flag antibodies. Actin served as loading control. A densitometric quantification of the relative expression levels is shown in the bar graph. (B) Gel filtration on Sephacryl S300. Extracts from 293fENL cells were separated by gel filtration on a precalibrated column as indicated. Individual eluate fractions were tested by immunoblotting with antibodies against ENL and flag. The elution profile of ENL from the parental HEK293 cells is shown in the bottom panel.
Figure 2
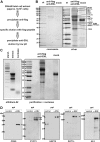
Purification of EAP. (A) Schematic flow chart of the purification procedure. (B) Composition of EAP. EAP was purified from approximately 5 × 108 293fENL cells. As a mock control, the same purification strategy was performed using unliganded agarose in place of flag-agarose for the first precipitation step (mock). Eluates were run on a 10% SDS-PAGE, stained with Coomassie (left panel) and subsequently with silver (right panel). The bands were excised, digested, and analyzed by mass spectrometry. All proteins that could be uniquely identified by more than 1 peptide and at least in 2 independent experiments are listed. Where individual band identities could be assigned, they are indicated in the figure. (C) Left panel shows nucleic acid content of cellular extracts used for EAP purification. Nucleic acids from cellular extracts used for EAP purification were isolated by phenol extraction, separated by agarose gel electrophoresis and stained by ethidium bromide. Samples shown are from untreated cellular extracts (extract) and from extracts after nuclease digestion (nuclease). Right panel shows purification of EAP from nuclease-treated cell extracts. EAP was precipitated as described in panel B from cellular extracts after exhaustive nuclease digestion. The figure shows a silver stained gel of EAP proteins in com-parison to a mock control as described in panel B. (D) Coimmunoprecipitation of major EAP components with endogenous ENL. Immunoprecipitations from HEK293 cell extracts were done either with agarose coupled to anti-ENL antibodies (α-ENL) or flag-agarose (control). Coprecipitating proteins were analyzed by immunoblot with antibodies against the pTEFb subunits CDK9, CYCT2, and HSP70, as well as with antibodies specific for DOT1L and AF4. The 2 splice variants of CYCT2 (CYCT2a, 2b) are indicated.
Figure 3
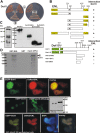
ENL interacts directly with Dot1l. (A) Example of a 2-hybrid experiment using full-length ENL as bait and full-length Dot1l as an interaction target. For comparison, yeast transformed with empty vectors (−) or with plasmids encoding the interacting proteins SNF1 and SNF4 (+) was plated alongside. Growth is shown on control and selective plates. (B) Structure function analysis to determine the ENL-Dot1l association interface. A series of ENL mutants was tested as bait for interaction with full-length Dot1l and vice versa to delineate the respective interaction domains in 2-hybrid experiments. Growth on selective medium is indicated with “+” or “−.” Conserved domains in ENL are labeled “YEATS” (a domain found in several other proteins associated with chromatin modification) and “hydp” (hydrophobic C-terminal domain that has been shown to mediate binding to CBX8). (C) Control for expression of Dot1l derivatives in yeast cells. The correct expression of the Dot1l mutants was checked by immunoblotting with a GAL4-activation domain–specific antibody. The lane designations correspond to the Dot1l mutants shown in panel B. The correct expression of the indicated set of ENL mutants has been published previously. (D) GST pull-down experiment. Beads loaded with purified GST or GST fused to full-length ENL were incubated with 35S-labeled Dot1l protein produced by in vitro transcription/translation. After washing, bound proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and visualized by autoradiography. (E) In vivo colocalization of ENL and Dot1l. Top row: vectors encoding fusions of EGFP with Dot1l and RFP with ENL were transfected into HEK293 cells. Fluorescent proteins were detected by microscopy in a nuclear speckled pattern. Photographs in the green and red channels and a software overlay are shown. Middle row: EGFP was fused with a mutant of Dot1l deleting amino acids 937 to 1095 within the ENL-binding domain. The protein was introduced into HEK293 cells (EGFP-Dot1lΔENLbdg), and the expression pattern was compared with wild-type EGFP-Dot1l (EGFP-Dot1l). Correct expression of the respective fusion proteins was controlled by immunoblotting with a GFP-specific antibody. Bottom row: colocalization of EGFP-Dot1l and RFP-ENL in Triton X-100–permeabilized cells to allow counterstaining with the DNA stain DAPI. All microphotographs were taken at room temperature in tissue culture medium with a Canon Coolpix 990 (Krefeld, Germany) electronic camera attached to a Zeiss Axioskop microscope (Oberkochen, Germany) with a Zeiss Neofluar 63×/1.25 NA objective and processed with Corel Draw software (Unterschleißheim, Germany).
Figure 4
Analysis of EAP histone methyltransferase and CDK kinase activities in vitro. (A) EAP contains a H3K79 dimethylating activity. Left panel: an EAP preparation or a mock control that was done with unliganded agarose as precipitation agent were incubated with core histones and methyl-3H-S-adenosylmethionine. The reaction products and a histone input sample (100% input) were separated on a 18% SDS gel, stained with Coomassie blue, and treated with a fluorographic enhancer solution before exposure to x-ray film (fluorography). Right panel: in an analogous experiment, core histones were incubated with EAP or a mock control as in panel A, and nonradioactive S-adenosylmethionine. After separation of the reaction products by 18% SDS-PAGE, the proteins were blotted and analyzed by immunodetection first with a H3K79 dimethyl–specific antibody and subsequently on the same membrane with a pan-H3–reactive antibody. As a further control, aliquots of the EAP and mock reactions were probed by Western blot for changes of dimethylation at H3K9. The densitometrically obtained concentration values are given underneath the respective lanes. (B) EAP has RNAPolII CTD kinase and pTEFb activity in vitro. Left panel: EAP works as RNAPolII CTD kinase in vitro. Recombinant GST–RNAPolII CTD fusion protein was incubated in the presence of γ32P ATP and either an EAP preparation or a mock control as described. Reactions were separated by SDS-PAGE followed by Coomassie staining and autoradiography. Right panel: EAP supplies pTEFb activity for in vitro transcription. Standard run-off transcription reactions in HeLa nuclear extracts were primed with a template allowing the transcription of the first 244 bp of the Hoxa9 coding sequence under control of the CMV immediate early promoter. The reaction was carried out with α32P-UTP to allow labeling of the transcript. The reaction products were phenolized and separated on a denaturing 6 M urea 6% PAA gel followed by autoradiography. The transcription assays were supplemented either with a mock precipitate as described, or with an EAP preparation. Whereas transcript levels were reduced by addition of the pTEFb inhibitor DRB in unsupplemented samples (left), the presence of EAP rescues a significant amount of transcriptional activity under the same conditions (right).
Figure 5
EAP function in vivo. (A) ENL is necessary for global H3K79 dimethylation. Endogenous ENL in HEK293 cells was knocked down by transfection with a vector encoding an ENL-specific shRNA. As control, a similar vector with a luciferase-specific shRNA insert was used. Extracts from cells transfected as indicated were subjected to immunoblotting to detect dimethyl-H3K79, ENL, total histone H3, and actin. A densitometric evaluation of 3 different experiments giving average values and standard deviations is shown in the bar graphs. (B) ENL is necessary for gene-specific H3K79 dimethylation. The distribution of dimethylated H3K79 across the HOXA9 coding region was detected by ChIP in control HEK293 cells (□; - - -) transfected with a luciferase-specific shRNA vector or in ENL knock-down cells (■; ▁) after treatment with anti ENL shRNA. As control, H3K4 di/trimethylation and H3K9 dimethylation was determined in analogous experiments. The localization of the primer pairs used for amplification of HOXA9 sequences is shown. An external primer pair amplifying untranscribed genomic sequence was used as control. Input and precipitate was quantified by quantitative PCR in triplicates. The percentage of specifically recovered material compared with input is plotted on the y-axis as average and standard deviation. (C) ENL knock-down affects global run-on elongation and endogenous as well as transient gene expression. Top panels: global elongation rates were measured by nuclear run-on experiments in isolated nuclei. Radioactivity incorporated into nascent RNA during the elongation process was measured in control cells (luciferase specific shRNA) and after ENL knock-down by ENL-specific shRNA as indicated. As control, the amount of stable 18S rRNA was determined in the same samples by quantitative real-time RT-PCR. Average and standard deviations of triplicate experiments are given. Bottom panels: Left panel shows labeled run-on RNA samples from ENL knock-down (sh) and control (luci) cells hybridized to cDNAs of the indicated genes. For normalization, 18S cDNA was included. Right panel shows total HOXA9-specific mRNA in ENL knock-down and control cells determined by qRT-PCR. (D) ENL knock-down affects transient transcription. Luciferase reporter experiments testing the response of 3 different promoters after transfection of increasing amounts of ENL specific shRNA vectors. Averages and standard deviations of triplicate experiments are plotted.
Figure 6
The transforming capability of an MLL-ENL fusion protein correlates with Dot1l binding. (A) Schematic depiction of alanine insertion mutations introduced into the ENL C-terminus. (B) Results of a serial replating assay with primary hematopoietic precursor cells isolated from bone marrow and transduced either with wild-type MLL-ENL, MLL-ENL mutant A, or MLL-ENL mutant B. Given are the average colony numbers including standard deviations in the third round of replating. Next to the bar graph, a representative example of stained colonies is shown. The insets show Western blot experiments detecting expression of the corresponding GAL4/MLL fusion proteins. (C) 2-hybrid test for interaction of ENL mutants A and B with full-length Dot1l and CBX8. Yeast was cotransformed with a bait plasmid containing mutants A and B as indicated and a Dot1l interaction target. Growth on adenine/histidine deficient selective medium is shown. (D) qRT-PCR analysis of MLL-ENL target gene expression. RNA was isolated from primary hematopoietic cells 1 week after transduction with retroviral constructs coding for wild-type MLL-ENL, mutant A, or mutant B, respectively. Relative RNA levels of the known MLL-ENL target genes Hoxa7, Hoxa9, and Meis1 were determined by qRT-PCR, normalized with respect to actin expression, and plotted with values for wild-type MLL-ENL transduced cells set arbitrarily to 1. Note that the results are displayed on a logarithmic scale. Average and standard deviation of triplicates are plotted.
Similar articles
- The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling.
Bitoun E, Oliver PL, Davies KE. Bitoun E, et al. Hum Mol Genet. 2007 Jan 1;16(1):92-106. doi: 10.1093/hmg/ddl444. Epub 2006 Nov 29. Hum Mol Genet. 2007. PMID: 17135274 - MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia.
Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. Monroe SC, et al. Exp Hematol. 2011 Jan;39(1):77-86.e1-5. doi: 10.1016/j.exphem.2010.09.003. Epub 2010 Sep 18. Exp Hematol. 2011. PMID: 20854876 Free PMC article. - The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin.
Zeisig DT, Bittner CB, Zeisig BB, García-Cuéllar MP, Hess JL, Slany RK. Zeisig DT, et al. Oncogene. 2005 Aug 18;24(35):5525-32. doi: 10.1038/sj.onc.1208699. Oncogene. 2005. PMID: 15856011 - The Intrinsically Disordered Proteins MLLT3 (AF9) and MLLT1 (ENL) - Multimodal Transcriptional Switches With Roles in Normal Hematopoiesis, MLL Fusion Leukemia, and Kidney Cancer.
Kabra A, Bushweller J. Kabra A, et al. J Mol Biol. 2022 Jan 15;434(1):167117. doi: 10.1016/j.jmb.2021.167117. Epub 2021 Jun 23. J Mol Biol. 2022. PMID: 34174329 Free PMC article. Review. - The molecular biology of mixed lineage leukemia.
Slany RK. Slany RK. Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review.
Cited by
- Transcription-associated histone modifications and cryptic transcription.
Smolle M, Workman JL. Smolle M, et al. Biochim Biophys Acta. 2013 Jan;1829(1):84-97. doi: 10.1016/j.bbagrm.2012.08.008. Epub 2012 Sep 7. Biochim Biophys Acta. 2013. PMID: 22982198 Free PMC article. Review. - TBP loading by AF4 through SL1 is the major rate-limiting step in MLL fusion-dependent transcription.
Okuda H, Takahashi S, Takaori-Kondo A, Yokoyama A. Okuda H, et al. Cell Cycle. 2016 Oct 17;15(20):2712-22. doi: 10.1080/15384101.2016.1222337. Epub 2016 Aug 26. Cell Cycle. 2016. PMID: 27564129 Free PMC article. - Inhibition of HIV-1 transcription and replication by a newly identified cyclin T1 splice variant.
Gao G, Wu X, Zhou J, He M, He JJ, Guo D. Gao G, et al. J Biol Chem. 2013 May 17;288(20):14297-14309. doi: 10.1074/jbc.M112.438465. Epub 2013 Apr 8. J Biol Chem. 2013. PMID: 23569210 Free PMC article. - Super-enhancers complexes zoom in transcription in cancer.
Wang M, Chen Q, Wang S, Xie H, Liu J, Huang R, Xiang Y, Jiang Y, Tian D, Bian E. Wang M, et al. J Exp Clin Cancer Res. 2023 Jul 28;42(1):183. doi: 10.1186/s13046-023-02763-5. J Exp Clin Cancer Res. 2023. PMID: 37501079 Free PMC article. Review. - Inhibition of histone methyltransferase EZH2 depletes leukemia stem cell of mixed lineage leukemia fusion leukemia through upregulation of p16.
Ueda K, Yoshimi A, Kagoya Y, Nishikawa S, Marquez VE, Nakagawa M, Kurokawa M. Ueda K, et al. Cancer Sci. 2014 May;105(5):512-9. doi: 10.1111/cas.12386. Epub 2014 Mar 30. Cancer Sci. 2014. PMID: 24612037 Free PMC article.
References
- Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program. 2004:80–97. - PubMed
- Hess JL. Mechanisms of transformation by MLL. Crit Rev Eukaryot Gene Expr. 2004;14:235–254. - PubMed
- Slany RK. When epigenetics kills: MLL fusion proteins in leukemia [review]. Hematol Oncol. 2005;23:1–9. - PubMed
- Schreiner SA, Garcia-Cuellar MP, Fey GH, Slany RK. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia. 1999;13:1525–1533. - PubMed
- So CW, Cleary ML. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood. 2003;101:633–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources