Determinants of targeting by endogenous and exogenous microRNAs and siRNAs - PubMed (original) (raw)
. 2007 Nov;13(11):1894-910.
doi: 10.1261/rna.768207. Epub 2007 Sep 13.
Affiliations
- PMID: 17872505
- PMCID: PMC2040081
- DOI: 10.1261/rna.768207
Determinants of targeting by endogenous and exogenous microRNAs and siRNAs
Cydney B Nielsen et al. RNA. 2007 Nov.
Abstract
Vertebrate mRNAs are frequently targeted for post-transcriptional repression by microRNAs (miRNAs) through mechanisms involving pairing of 3' UTR seed matches to bases at the 5' end of miRNAs. Through analysis of expression array data following miRNA or siRNA overexpression or inhibition, we found that mRNA fold change increases multiplicatively (i.e., log-additively) with seed match count and that a single 8 mer seed match mediates down-regulation comparable to two 7 mer seed matches. We identified several targeting determinants that enhance seed match-associated mRNA repression, including the presence of adenosine opposite miRNA base 1 and of adenosine or uridine opposite miRNA base 9, independent of complementarity to the siRNA/miRNA. Increased sequence conservation in the approximately 50 bases 5' and 3' of the seed match and increased AU content 3' of the seed match were each independently associated with increased mRNA down-regulation. All of these determinants are enriched in the vicinity of conserved miRNA seed matches, supporting their activity in endogenous miRNA targeting. Together, our results enable improved siRNA off-target prediction, allow integrated ranking of conserved and nonconserved miRNA targets, and show that targeting by endogenous and exogenous miRNAs/siRNAs involves similar or identical determinants.
Figures
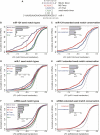
FIGURE 1.
Effects of seed match type and conservation on mRNA repression for miRNAs and siRNAs. (A) Seed match types and numbering system, illustrated for miR-1. Positions in the miRNA are numbered 5′-3′. (Seed match 6 mer) WC inverse complement of miRNA bases 2–7; (A1) presence of adenosine opposite miRNA base 1; (M8) WC match to miRNA base 8. (B) CDFs (cumulative distribution functions) of LFCs (log2 fold change) for mRNAs containing indicated miR-124 seed match types (colored lines and labels) or no miR-124 seed matches (gray line) following transfection of miR-124. (Solid vertical gray line) The LFC above which 97.5% of the no-seed-match mRNA set falls. (Inset bar plot) nLFC (normalized log2 fold change) values for each seed match type with error bars indicating standard error. Data for panels B_–_E are from Lim et al. (2005). Plots include only mRNAs containing exactly one miR-124 seed match, and thus the seed match type sets are mutually exclusive. The distribution of mRNA expression values did not differ significantly between seed match type sets (P > 0.05 by rank sum test). All seed match types except the 6 mer have distributions significantly different from the no-seed-match class (P < 0.005 by rank sum test). (_C_) CDFs of LFCs for mutually exclusive mRNA sets containing conserved (red) or nonconserved (blue) extended seed matches to miR-124, or no seed matches (gray); the conserved and nonconserved sets are significantly different (_P_ < 0.001 by rank sum test). The “nonconserved” mRNA set contains exclusively nonconserved seed matches; the “conserved” mRNAs may also contain nonconserved seed matches. The nonconserved set was sampled to match the conserved set in seed match type and count, overall UTR conservation, and initial mRNA expression level (Supplemental Fig. S2). (_D_) Same as _B_ for miR-1. All seed match type classes are significantly different from the no-seed-match class (_P_ < 10−4 by rank sum test). (_E_) Same as _C_ for miR-1. The CDFs of conserved and nonconserved mRNA sets are significantly different (_P_ < 0.01 by rank sum test). (_F_) Same as _B_ for pooled set of 33 “effective” siRNAs that begin with non-U bases (Supplemental Material). Additional seed match classes containing M1 are shown (triangles). All seed match types have distributions significantly different from the no-seed-match class (_P_ < 10−14 by rank sum test). (_G_) Same analysis and controls as _C_ for pooled set of siRNAs. The CDFs of conserved and nonconserved mRNA sets are not significantly different (_P_ > 0.05 by rank sum test). See Supplemental Table S1 for additional statistics.
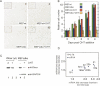
FIGURE 2.
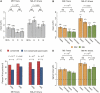
Characterization of conditional Dicer knockout (CDKO) mouse embryonic fibroblasts (MEFs). (A) Wild-type (wt) and CDKO MEFs are shown, untreated (left panels) or 4 d after ortho hydroxy tamoxifen (OHT) treatment (right panels). Cells were stained for LacZ, and the percentage of LacZ-positive cells is shown (upper right). (B) Proliferation of wild-type and CDKO MEFs, untreated or following addition of OHT. Error bars represent standard deviation of three independent counts. (C) Western analysis of MEFcdko cells, untreated (lane 4) or after OHT addition (lane 5), showing a three- to fourfold reduction in Dicer protein levels following OHT addition. GAPDH is a loading control. Westerns using different concentrations of recombinant Dicer protein are shown as a positive control (lanes 1,2). (D) Microarray hybridization intensity change and expression level change measured by Northern analysis (log scale, both axes) for eight miRNAs (miR-21, miR-22, miR-23b, miR-34a, miR-92, miR-191, miR-199a*, and miR-200b).
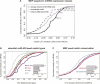
FIGURE 3.
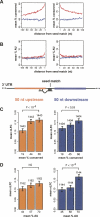
mRNA derepression following Dicer knockout varies with seed match type and conservation status. (A) CDFs of median LFC for three classes of mRNA sets. The expression classes were: (1) mRNA sets containing extended seed matches to the 80 miRNA families whose expression was detected above background by microarray (black curve, selected miRNA family names shown); (2) mRNA sets containing extended seed matches to the 50 miRNA families that were not detectably expressed (gray curve); (3) randomly selected mRNA sets (dotted line). Distributions of detected and nondetected sets are significantly different (P < 0.01 by rank sum test), but distributions of nondetected and random mRNA sets are not. (B) CDFs of LFCs for mRNAs containing the indicated miR-430 seed match types—or no miR-430 seed matches (gray curve)—for MZdicer zebrafish embryo data (Giraldez et al. 2006); only mRNAs with exactly one miR-430 seed match were included (seed match type mRNA sets are mutually exclusive). All seed match type LFC distributions differed significantly from the no-seed-match class (P < 10−7 by rank sum test), and the pooled extended seed match types differed from the 6 mer class (P < 0.01 by rank sum test). (Solid vertical gray line) The LFC below which 97.5% of the no-seed-match mRNA set falls. (Inset bar plot) LFC values for each seed match type, with error bars indicating standard error. (C) CDFs of LFCs for mRNAs containing conserved (red) or exclusively nonconserved (blue) extended seed matches, or no seed matches (gray) to the set of 31 “responsive” miRNAs in the CDKO MEF experiment (Supplemental Material). Seed match count, overall UTR conservation, and mRNA expression level were controlled between the sets. The distributions of mRNA with conserved and nonconserved seed matches are significantly different (P < 0.05 by rank sum test). See Supplemental Table S6 for additional statistics.
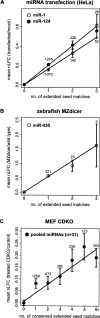
FIGURE 4.
mRNA fold change increases multiplicatively with extended seed match count. (A) For miR-1 (open circles) and miR-124 (solid black circles), the total number of extended seed matches was enumerated for each mRNA, and the mean nLFCs in the Lim transfection experiments were determined for sets of mRNAs grouped by seed match count (set sizes indicated above or below points). (Solid lines) Least squares fit for the whole data set. Error bars correspond to standard error. For each of these plots, the proportions of different seed match types for different seed match counts remained fairly constant. (B) Same as A for miR-430 extended seed match counts following Dicer knockout in zebrafish embryos (Giraldez et al. 2006). (C) Same as A for conserved extended seed match counts to 31 “responsive” mouse miRNAs (see Supplemental Material) in CDKO MEFs.
FIGURE 5.
Increased down-regulation of mRNAs with adenosine or uridine at position t9. (A) Mean nLFC for mRNAs containing the indicated nucleotide at position t9 flanking siRNA M8 7 mer and M8-A1 8 mer (rank sum test _P_-values; NS = not significant at _P_-value cutoff 0.05). Error bars indicate standard error, and the numbers of mRNAs are indicated above the bars. Each mRNA contained exactly one seed match to any given siRNA (i.e., t9 sets are mutually exclusive), and mRNAs in each of the four t9 sets were controlled for 3′ UTR GC content. Other variables, such as mRNA expression, 3′ UTR conservation, or m9 composition, did not differ significantly between t9 sets. (B) Same as A, but reclassifying the controlled mRNA sets by whether the t9 base pairs with the siRNA m9 (match) or not (mismatch). (C) Enrichment of t9W nucleotides flanking conserved versus nonconserved miRNA M8 7 mer and M8-A1 8 mer in human 3′ UTRs (χ2 test _P_-values). The miRNA set consisted of conserved human miRNAs used for target prediction by Lewis et al. (2005) after removal of miRNAs with common m2–m8 seed regions but different m9 nucleotides, and pairs of miRNAs in the same superfamily. The nonconserved seed matches were sampled to match the seed match type, miRNA, and overall UTR CG content of the conserved set. (D) Mean signal:noise ratios for M8 7 mer and M8-A1 8 mer with t9W or t9S in match and mismatch configurations based on cohorts of control oligonucleotides (Lewis et al. 2005) matched for both count and exact CG content (error bars indicate standard deviation based on 14 control cohorts). (Dashed line) Baseline S:N value of 1. _P_-values based on Wilcoxon rank sum tests between indicated sets (NS = not significant at _P_-value cutoff 0.05).
FIGURE 6.
siRNA-directed mRNA repression is enhanced by local conservation and AU content. (A) Mean percent conservation at UTR positions within 100 bp 5′ (left) and 3′ (right) of conserved (red) or nonconserved (blue) extended seed matches to the set of conserved vertebrate miRNAs used in Lewis et al. (2005); overall UTR conservation was controlled for in the comparison of conserved and nonconserved seed matches. The average conservation differs significantly for the 100 bases 5′ and 3′ of the seed match (P < 10−200 by rank sum test for both). (B) Mean AU content (sets controlled for UTR AU content); average AU content is significantly different both 5′ and 3′ of the seed match (P < 10−18, P < 10−30), respectively. (C) Mean nLFC for three equal-sized mRNA sets binned by percent conserved positions (in HMRD) in the 50-nt region immediately 5′ (orange) or 3′ (purple) of siRNA seed matches (5′ region ends at position t10; 3′ region begins one base 3′ of position t1) for mRNAs containing single extended seed match to the relevant siRNA. Bars indicate standard error of the mean. Set size and mean percent conservation for each set are reported above and below each bar, respectively. _P_-values are for two-sided rank sum tests between the first and third bins. For both 5′ and 3′ conservation, the three bins have been sampled such that their distributions of overall UTR conservation, 5′ (or 3′) AU content, overall UTR AU content, seed match type, and initial expression level are not significantly different (P ≥ 0.05) (Supplemental Fig. S9). (D) Same as C, but with UTRs binned by AU content in the same 50-nt regions. Bins are sampled to control for UTR AU content, 5′ (or 3′) conservation, overall UTR conservation, seed match type, and initial expression level (NS = not significant at _P_-value cutoff 0.05).
FIGURE 7.
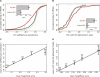
TargetRank scoring separates strongly and weakly down-regulated mRNAs. (A) CDFs of LFCs (as in Fig. 1) for the top 20% (red) and bottom 20% (green) of mRNAs to the relevant siRNA in the test set of eight randomly chosen siRNA transfection experiments ranked by TargetRank score. Only expressed mRNAs containing exactly one 7 mer seed match and no other seed matches of any type were used. For reference, the CDF for mRNAs lacking seed matches to the relevant siRNAs is shown (gray). (Solid vertical gray line) The LFC above which 97.5% of the no-seed-match mRNA set falls. (Inset bar plot) See Fig. 1. See Supplemental Table S7 for additional statistics. (B) Same as A, but for miR-155 knockout T cell data from Rodriguez et al. (2007). (C) All mRNAs containing seed matches (of any type or count) to the relevant siRNA in the eight test siRNA transfection experiments (same sets as in A) were scored using TargetRank. The mean TargetRank score and mean nLFC are plotted with standard error bars for mRNAs sets binned by TargetRank score (mRNA set sizes indicated above points). Line corresponds to least squares fit for entire data set (ANOVA P < 10−100); r = 0.23 (Pearson correlation). (D) Same as C, but for miR-155 knockout T cell data from (Rodriguez et al. 2007). Line corresponds to least squares fit for entire data set (ANOVA P < 10−18); r = 0.24 (Pearson correlation).
Similar articles
- siRNAs can function as miRNAs.
Doench JG, Petersen CP, Sharp PA. Doench JG, et al. Genes Dev. 2003 Feb 15;17(4):438-42. doi: 10.1101/gad.1064703. Genes Dev. 2003. PMID: 12600936 Free PMC article. - HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR.
Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. Klase Z, et al. BMC Mol Biol. 2007 Jul 30;8:63. doi: 10.1186/1471-2199-8-63. BMC Mol Biol. 2007. PMID: 17663774 Free PMC article. - MicroRNA targeting specificity in mammals: determinants beyond seed pairing.
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Grimson A, et al. Mol Cell. 2007 Jul 6;27(1):91-105. doi: 10.1016/j.molcel.2007.06.017. Mol Cell. 2007. PMID: 17612493 Free PMC article. - Dicer cuts the kidney.
Ho JJ, Marsden PA. Ho JJ, et al. J Am Soc Nephrol. 2008 Nov;19(11):2043-6. doi: 10.1681/ASN.2008090986. Epub 2008 Oct 15. J Am Soc Nephrol. 2008. PMID: 18923053 Review. No abstract available. - Is the Efficiency of RNA Silencing Evolutionarily Regulated?
Ui-Tei K. Ui-Tei K. Int J Mol Sci. 2016 May 12;17(5):719. doi: 10.3390/ijms17050719. Int J Mol Sci. 2016. PMID: 27187367 Free PMC article. Review.
Cited by
- Stage-specific expression patterns and co-targeting relationships among miRNAs in the developing mouse cerebral cortex.
Todorov H, Weißbach S, Schlichtholz L, Mueller H, Hartwich D, Gerber S, Winter J. Todorov H, et al. Commun Biol. 2024 Oct 22;7(1):1366. doi: 10.1038/s42003-024-07092-7. Commun Biol. 2024. PMID: 39433948 Free PMC article. - Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis.
Yadav V, Singh T, Sharma D, Garg VK, Chakraborty P, Ghatak S, Satapathy SR. Yadav V, et al. Cancers (Basel). 2024 Sep 18;16(18):3183. doi: 10.3390/cancers16183183. Cancers (Basel). 2024. PMID: 39335155 Free PMC article. Review. - A high-resolution map of functional miR-181 response elements in the thymus reveals the role of coding sequence targeting and an alternative seed match.
Verheyden NA, Klostermann M, Brüggemann M, Steede HM, Scholz A, Amr S, Lichtenthaeler C, Münch C, Schmid T, Zarnack K, Krueger A. Verheyden NA, et al. Nucleic Acids Res. 2024 Aug 12;52(14):8515-8533. doi: 10.1093/nar/gkae416. Nucleic Acids Res. 2024. PMID: 38783381 Free PMC article. - Advanced computational predictive models of miRNA-mRNA interaction efficiency.
Bader S, Tuller T. Bader S, et al. Comput Struct Biotechnol J. 2024 Apr 19;23:1740-1754. doi: 10.1016/j.csbj.2024.04.015. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38689718 Free PMC article. - Experimental capture of miRNA targetomes: disease-specific 3'UTR library-based miRNA targetomics for Parkinson's disease.
Hart M, Kern F, Fecher-Trost C, Krammes L, Aparicio E, Engel A, Hirsch P, Wagner V, Keller V, Schmartz GP, Rheinheimer S, Diener C, Fischer U, Mayer J, Meyer MR, Flockerzi V, Keller A, Meese E. Hart M, et al. Exp Mol Med. 2024 Apr;56(4):935-945. doi: 10.1038/s12276-024-01202-5. Epub 2024 Apr 1. Exp Mol Med. 2024. PMID: 38556547 Free PMC article.
References
- Abbondanzo, S.J., Gadi, I., Stewart, C.L. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225:803–823. - PubMed
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
- Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous