In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis - PubMed (original) (raw)
In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis
Hirotaka Yoshioka et al. Mol Cell Biol. 2007 Nov.
Abstract
Transcription of the testis-specific Pgk2 gene is selectively activated in primary spermatocytes to provide a source of phosphoglycerate kinase that is critical to normal motility and fertility of mammalian spermatozoa. We examined dynamic changes in protein-DNA interactions at the Pgk2 gene promoter during murine spermatogenesis in vivo by performing genomic footprinting and chromatin immunoprecipitation assays with enriched populations of murine spermatogenic cells at stages prior to, during, and following transcription of this gene. We found that genes encoding the testis-specific homeodomain factor PBX4 and its coactivator, PREP1, are expressed in patterns that mirror expression of the Pgk2 gene and that these factors become bound to the Pgk2 enhancer in cells in which this gene is actively expressed. We therefore suggest that these factors, along with CREM and SP3, direct stage- and cell type-specific transcription of the Pgk2 gene during spermatogenesis. We propose that binding of PBX4, plus its coactivator PREP1, is a rate-limiting step leading to the initiation of tissue-specific transcription of the Pgk2 gene. This study provides insight into the developmentally dynamic establishment of tissue-specific protein-DNA interactions in vivo. It also allows us to speculate about the events that led to tissue-specific regulation of the Pgk2 gene during mammalian evolution.
Figures
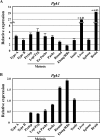
FIG. 1.
Quantitative RT-PCR analysis of Pgk gene expression during spermatogenesis. Expression of the Pgk1 (A) and Pgk2 (B) genes during spermatogenesis and in representative somatic tissues was quantified by real-time RT-PCR. The relative expression of target mRNA was calculated from the target CT values and β-actin mRNA CT values, using the standard curve method. Each error bar indicates standard error of the mean. Populations of the specific spermatogenic cell types examined included the following: Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids; Elong/RBs, elongated spermatids plus residual bodies.
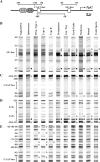
FIG. 2.
In vivo footprinting of the Pgk2 promoter during spermatogenesis in the mouse. (A) Schematic representation of putative regulatory elements in the Pgk2 gene promoter. The transcription start site is represented as a bent arrow. The core promoter includes a GC box and a CAAT box (open boxes). Immediately upstream from the core promoter is enhancer region E1/E2 and the E3/E4 subregions that have been shown to bind up to four anonymous factors. These sites are represented, respectively, as oval light and dark gray boxes. The location of amplimers analyzed by LM-PCR for in vivo footprinting are depicted as horizontal lines with numbers representing nucleotide positions relative to the transcription start site. Amplimers shown above and below the line represent analysis of upper and lower DNA strands, respectively. (B through E) DMS modification and LM-PCR were used to in vivo footprint the core promoter and enhancer regions of the Pgk2 promoter in purified populations of mouse spermatogenic cells. Open circles adjacent to bands represent protected cleavage sites, whereas filled circles represent enhanced cleavage sites. Numbers represent nucleotide positions relative to the transcriptional initiation site. Vertical lines represent the location of known or putative factor-binding elements. Prim. type A, primitive type A spermatogonia; type A, type A spermatogonia; type B, type B spermatogonia; Lep/Zyg, leptotene plus zygotene spermatocytes; early pachs, early (juvenile) pachytene spermatocytes; pachytene, pachytene spermatocytes; round, round spermatids; testis sperm, testicular sperm.
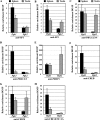
FIG. 3.
In vivo binding of transcription factors to the Pgk1 and Pgk2 promoters. ChIP analyses were performed with testicular germ cells and splenocytes, using antibodies specific for SP1 (A), SP3 (B), PBX1/2/3/4 (C), PBX1/2/3 (D), PREP1 (E), CREB (F), CREM (G), and CBF-B/NF-YA (H). Precipitated DNA was subjected to real-time PCR with the use of specific primers for the Pgk1 and Pgk2 promoters (see below), and relative enrichment of amplified regions in immunoprecipitated DNA (bound fractions; BO) was determined and compared to that in input (IP) DNA. All results were normalized to values for normal rabbit IgG, with that level arbitrarily set to 1.0. Each error bar indicates standard error of the mean. All PCR primers used are listed in Table 2. The PCR primer set named “Pgk1 CAAT box” was used to analyze the binding of all transcription factors examined in the Pgk1 promoter. This primer set amplified the “NF1-like region” (from position −180 to −77) (14, 47), including the consensus binding site for PBX factors and the CAAT box element (the binding site for CBF-B/NF-YA) in the Pgk1 promoter. Because of the size of the sonication fragments, amplification of this region was also used to examine the binding of SP factors to the nearby GC boxes at positions −66 and −254 in the Pgk1 promoter. Similarly, the PCR primer sets named “Pgk2 E1-E4,” “Pgk2 CAAT box,” and “Pgk2 GC box” (Table 2) were used to selectively amplify specific regions in the Pgk2 promoter, including a region containing the E1/E2-plus-E3/E4 enhancer sites, a region containing the CAAT box, and a region containing the GC box, respectively. The “Pgk2 GC box” primer set was also used to analyze the binding of CREB and/or CREM to the putative CRE site (+333). Deduction of specificity of binding of each factor to each element was based on the fact that, except where noted otherwise, each putative binding element represented the only candidate consensus binding sequence for each factor within at least 1 kb in each direction in each promoter.
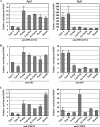
FIG. 4.
Developmental stage- and cell type-specific recruitment of transcription factors to the Pgk1 and Pgk2 promoters. ChIP analyses were performed with relatively pure populations of specific spermatogenic cell types prior to, during, and following the period of active transcription of the Pgk2 gene, using antibodies specific for PBX1/2/3/4 (A), SP3 (B), and CREM (C). Precipitated DNA was subjected to real-time PCR with the use of specific primers for the Pgk2 (left side panels) and Pgk1 (right side panels) promoters, and enrichment of immunoprecipitated DNA (bound fractions, BO) relative to input (IP) DNA was calculated in each case. All results were normalized to values for normal rabbit IgG, with that level arbitrarily set to 1.0. Each error bar indicates standard error of the mean. Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids.
FIG. 5.
qRT-PCR analysis of expression during spermatogenesis of genes encoding PBX and MEIS factors. Expression of different members of the Pbx family of genes, Pbx1a (A), Pbx1b (B), Pbx2 (C), Pbx3 (D), and Pbx4 (E), and the Meis family of genes, Meis1 (F), Meis2 (G), Meis3 (H), Prep1 (I), and Prep2 (J), was quantitated by real-time RT-PCR. The relative expression of each target mRNA was calculated from the target CT values and the 18S rRNA CT values, using the standard curve method. Each error bar indicates standard error of the mean. Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids; Elong/RBs, elongated spermatids plus residual bodies.
FIG. 6.
Pgk promoter sequences in mammals. Comparisons are shown among promoter sequences for the Pgk1 (A) and Pgk2 (B) genes from various mammalian species. Alignment of sequences upstream from the translational start codon (ATG, dashed rectangle) of each Pgk gene was done using CLUSTALW (
). Identical nucleotides are represented by asterisks, and dashes indicate a gap. The numbers to the right of each sequence are relative to the transcription start site, which is designated +1. Highly conserved sequences for which potential binding functions are known are surrounded by rectangles. (C) An alignment is shown of the NF1-like region from the human and mouse Pgk1 genes and the E3/E4 region from the human and mouse Pgk2 genes and the potential relationship among these regions. Asterisks indicate sequence identity, dashes indicate a lack of identity, and vertical lines indicate partial identity.
Similar articles
- Sequence-specific promoter elements regulate temporal-specific changes in chromatin required for testis-specific activation of the Pgk2 gene.
Yang Z, Yoshioka H, McCarrey JR. Yang Z, et al. Reproduction. 2013 Oct 4;146(5):501-16. doi: 10.1530/REP-13-0311. Print 2013. Reproduction. 2013. PMID: 24000349 Free PMC article. - Epigenetic regulation of testis-specific gene expression.
McCarrey JR, Geyer CB, Yoshioka H. McCarrey JR, et al. Ann N Y Acad Sci. 2005 Dec;1061:226-42. doi: 10.1196/annals.1336.025. Ann N Y Acad Sci. 2005. PMID: 16467272 - Ontogeny of a demethylation domain and its relationship to activation of tissue-specific transcription.
Geyer CB, Kiefer CM, Yang TP, McCarrey JR. Geyer CB, et al. Biol Reprod. 2004 Sep;71(3):837-44. doi: 10.1095/biolreprod.104.028969. Epub 2004 May 12. Biol Reprod. 2004. PMID: 15140797 - Testis-specific transcription mechanisms promoting male germ-cell differentiation.
Kimmins S, Kotaja N, Davidson I, Sassone-Corsi P. Kimmins S, et al. Reproduction. 2004 Jul;128(1):5-12. doi: 10.1530/rep.1.00170. Reproduction. 2004. PMID: 15232059 Review. - CREM: a master-switch in the transcriptional response to cAMP.
Lamas M, Monaco L, Zazopoulos E, Lalli E, Tamai K, Penna L, Mazzucchelli C, Nantel F, Foulkes NS, Sassone-Corsi P. Lamas M, et al. Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):561-7. doi: 10.1098/rstb.1996.0055. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735279 Review.
Cited by
- A cytoplasmic variant of the KH-type splicing regulatory protein serves as a decay-promoting factor for phosphoglycerate kinase 2 mRNA in murine male germ cells.
Xu M, McCarrey JR, Hecht NB. Xu M, et al. Nucleic Acids Res. 2008 Dec;36(22):7157-67. doi: 10.1093/nar/gkn800. Epub 2008 Nov 10. Nucleic Acids Res. 2008. PMID: 19015122 Free PMC article. - Proteomic biomarkers in seminal plasma as predictors of reproductive potential in azoospermic men.
Fietz D, Sgaier R, O'Donnell L, Stanton PG, Dagley LF, Webb AI, Schuppe HC, Diemer T, Pilatz A. Fietz D, et al. Front Endocrinol (Lausanne). 2024 Apr 9;15:1327800. doi: 10.3389/fendo.2024.1327800. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38654926 Free PMC article. - Non-Invasive Diagnostics of Male Spermatogenesis from Seminal Plasma: Seminal Proteins.
Ješeta M, Pospíšilová A, Mekiňová L, Franzová K, Ventruba P, Lousová E, Kempisty B, Oždian T, Žáková J, Crha I. Ješeta M, et al. Diagnostics (Basel). 2023 Jul 25;13(15):2468. doi: 10.3390/diagnostics13152468. Diagnostics (Basel). 2023. PMID: 37568830 Free PMC article. Review. - RNA-based gene duplication: mechanistic and evolutionary insights.
Kaessmann H, Vinckenbosch N, Long M. Kaessmann H, et al. Nat Rev Genet. 2009 Jan;10(1):19-31. doi: 10.1038/nrg2487. Nat Rev Genet. 2009. PMID: 19030023 Free PMC article. Review. - Expression patterns of SP1 and SP3 during mouse spermatogenesis: SP1 down-regulation correlates with two successive promoter changes and translationally compromised transcripts.
Ma W, Horvath GC, Kistler MK, Kistler WS. Ma W, et al. Biol Reprod. 2008 Aug;79(2):289-300. doi: 10.1095/biolreprod.107.067082. Epub 2008 Apr 16. Biol Reprod. 2008. PMID: 18417714 Free PMC article.
References
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. - PubMed
- Almstrup, K., J. E. Nielsen, M. A. Hansen, M. Tanaka, N. E. Skakkebaek, and H. Leffers. 2004. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol. Reprod. 70:1751-1761. - PubMed
- Ariel, M., H. Cedar, and J. McCarrey. 1994. Developmental changes in methylation of spermatogenesis-specific genes include reprogramming in the epididymis. Nat. Genet. 7:59-63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases