SmedGD: the Schmidtea mediterranea genome database - PubMed (original) (raw)
. 2008 Jan;36(Database issue):D599-606.
doi: 10.1093/nar/gkm684. Epub 2007 Sep 18.
Affiliations
- PMID: 17881371
- PMCID: PMC2238899
- DOI: 10.1093/nar/gkm684
SmedGD: the Schmidtea mediterranea genome database
Sofia M C Robb et al. Nucleic Acids Res. 2008 Jan.
Abstract
The planarian Schmidtea mediterranea is rapidly emerging as a model organism for the study of regeneration, tissue homeostasis and stem cell biology. The recent sequencing, assembly and annotation of its genome are expected to further buoy the biomedical importance of this organism. In order to make the extensive data associated with the genome sequence accessible to the biomedical and planarian communities, we have created the Schmidtea mediterranea Genome Database (SmedGD). SmedGD integrates in a single web-accessible portal all available data associated with the planarian genome, including predicted and annotated genes, ESTs, protein homologies, gene expression patterns and RNAi phenotypes. Moreover, SmedGD was designed using tools provided by the Generic Model Organism Database (GMOD) project, thus making its data structure compatible with other model organism databases. Because of the unique phylogenetic position of planarians, SmedGD (http://smedgd.neuro.utah.edu) will prove useful not only to the planarian research community, but also to those engaged in developmental and evolutionary biology, comparative genomics, stem cell research and regeneration.
Figures
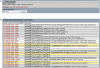
Figure 1.
Screen capture of SmedGD displaying genomic contig v31.019651. This contig has only one predicted gene, which has 5 exons and a 3′UTR. The tracks displayed include the gene model, its corresponding predicted transcript, and the relevant biological evidence associated with this model (see text for detailed explanations of each track). From this data, users can see the details of the gene model and its evidence (all of the predicted exons are supported by EST and protein evidence), and that the gene is likely coding for a histone deacetylase. Double-stranded RNA has been used to silence this gene and the resulting phenotypes are listed. cDNA Microarray data is not yet available, but a sample of how this information will be viewed is presented. An arrow pointing down indicates down-regulation of the gene in the experimental group.
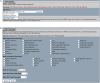
Figure 2.
(A) The ‘Protein Homology’ Search interface. In this example, the search term ‘piwi-like’ is being submitted. Each of the hits from SwissProt, SMART, PFAM and the species-specific databases are searched for the user query term. (B) Results of the search are sorted by genomic contig and location. When more than one result is found on one contig, the matches are grouped and the background will be similarly colored. When there is more than one protein match for one genomic location, it is often due to this sequence matching more than one database. When there is only one result per contig the background is colored white. The contig and location are hyperlinked to the genome browser for further inspection.
Figure 3.
(A) ‘Gene Ontology’ Search interface. Users can search for terms, such as ‘stem cell’ in one of three Gene Ontology (GO) categories (e.g. Biological Process). Any protein homology hit to the genome that had GO terms associated with it will be searched and the corresponding genomic contig and location will be returned in a fashion similar to the ‘Protein Homology’ results page. (B) The ‘RNAi Phenotype’ search interface is used by selecting phenotypes from the five categories listed. More information about these categories and phenotypes can be found in (3). The selections are additive, therefore each of the records returned will contain all of the chosen phenotypes.
Similar articles
- SmedGD 2.0: The Schmidtea mediterranea genome database.
Robb SM, Gotting K, Ross E, Sánchez Alvarado A. Robb SM, et al. Genesis. 2015 Aug;53(8):535-46. doi: 10.1002/dvg.22872. Epub 2015 Jul 17. Genesis. 2015. PMID: 26138588 Free PMC article. - The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain.
Zayas RM, Hernández A, Habermann B, Wang Y, Stary JM, Newmark PA. Zayas RM, et al. Proc Natl Acad Sci U S A. 2005 Dec 20;102(51):18491-6. doi: 10.1073/pnas.0509507102. Epub 2005 Dec 12. Proc Natl Acad Sci U S A. 2005. PMID: 16344473 Free PMC article. - Smed454 dataset: unravelling the transcriptome of Schmidtea mediterranea.
Abril JF, Cebrià F, Rodríguez-Esteban G, Horn T, Fraguas S, Calvo B, Bartscherer K, Saló E. Abril JF, et al. BMC Genomics. 2010 Dec 31;11:731. doi: 10.1186/1471-2164-11-731. BMC Genomics. 2010. PMID: 21194483 Free PMC article. - [Regeneration of planarians: experimental object].
Sheĭman IM, Kreshchenko ID. Sheĭman IM, et al. Ontogenez. 2015 Jan-Feb;46(1):3-12. Ontogenez. 2015. PMID: 25898529 Review. Russian. - [Molecular approach to planarian stem cell system].
Agata K. Agata K. Tanpakushitsu Kakusan Koso. 2005 May;50(6 Suppl):706-10. Tanpakushitsu Kakusan Koso. 2005. PMID: 15926503 Review. Japanese. No abstract available.
Cited by
- HOX gene complement and expression in the planarian Schmidtea mediterranea.
Currie KW, Brown DD, Zhu S, Xu C, Voisin V, Bader GD, Pearson BJ. Currie KW, et al. Evodevo. 2016 Mar 30;7:7. doi: 10.1186/s13227-016-0044-8. eCollection 2016. Evodevo. 2016. PMID: 27034770 Free PMC article. - Collagen IV differentially regulates planarian stem cell potency and lineage progression.
Chan A, Ma S, Pearson BJ, Chan D. Chan A, et al. Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2021251118. doi: 10.1073/pnas.2021251118. Proc Natl Acad Sci U S A. 2021. PMID: 33859045 Free PMC article. - Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea.
Resch AM, Palakodeti D, Lu YC, Horowitz M, Graveley BR. Resch AM, et al. PLoS One. 2012;7(4):e34447. doi: 10.1371/journal.pone.0034447. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496805 Free PMC article. - PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells.
Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA. Rouhana L, et al. Development. 2012 Mar;139(6):1083-94. doi: 10.1242/dev.076182. Epub 2012 Feb 8. Development. 2012. PMID: 22318224 Free PMC article. - Helminth genomics: The implications for human health.
Brindley PJ, Mitreva M, Ghedin E, Lustigman S. Brindley PJ, et al. PLoS Negl Trop Dis. 2009 Oct 26;3(10):e538. doi: 10.1371/journal.pntd.0000538. PLoS Negl Trop Dis. 2009. PMID: 19855829 Free PMC article. Review.
References
- Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. - PubMed
- Sánchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124:241–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources