Short pyrosequencing reads suffice for accurate microbial community analysis - PubMed (original) (raw)
Short pyrosequencing reads suffice for accurate microbial community analysis
Zongzhi Liu et al. Nucleic Acids Res. 2007.
Abstract
Pyrosequencing technology allows us to characterize microbial communities using 16S ribosomal RNA (rRNA) sequences orders of magnitude faster and more cheaply than has previously been possible. However, results from different studies using pyrosequencing and traditional sequencing are often difficult to compare, because amplicons covering different regions of the rRNA might yield different conclusions. We used sequences from over 200 globally dispersed environments to test whether studies that used similar primers clustered together mistakenly, without regard to environment. We then tested whether primer choice affects sequence-based community analyses using UniFrac, our recently-developed method for comparing microbial communities. We performed three tests of primer effects. We tested whether different simulated amplicons generated the same UniFrac clustering results as near-full-length sequences for three recent large-scale studies of microbial communities in the mouse and human gut, and the Guerrero Negro microbial mat. We then repeated this analysis for short sequences (100-, 150-, 200- and 250-base reads) resembling those produced by pyrosequencing. The results show that sequencing effort is best focused on gathering more short sequences rather than fewer longer ones, provided that the primers are chosen wisely, and that community comparison methods such as UniFrac are surprisingly robust to variation in the region sequenced.
Figures
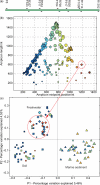
Figure 1.
16S community samples from a broad range of physical environments cluster by environment type, not by primers. (a) Popular sequencing primers, as shown in the European rRNA database. The concern is that sequences amplified using the same primer pair might artifactually cluster together, even if the microbial communities differ, due to primer bias. (b) Distribution of community samples according to midpoint and length of amplicon. Symbol indicates environment type (squares = soil, triangles = marine sediment, diamonds = fresh water, circles = other environments), size indicates length of amplicon (larger symbols indicate longer amplicons) and color spectrum indicates position of midpoint in the sequence (blue → red = start → end of sequence). (c) Distribution of community samples in UniFrac principal coordinates anaylsis (PCoA), colors and symbols same as in (b) above. Samples clearly cluster by environment type, rather than by amplicon, as symbols of the same color and shape are found in each of the environment type clusters. Circles in (b) and (c) show a single point on the primer length graph split into several related but distinct samples on the environment graph (six from different rivers, two from different lakes).
Figure 2.
UniFrac clustering with artificially shortened amplicons tends to recapture the same patterns as the full-length sequences. (a) Primer sequences as in Figure 1a, showing the artificial amplicons that were obtained by clipping the sequences using each primer pair. Sequences were truncated at positions 83 and 1326 (relative to the E. coli sequence) because this was the limit of the amplified region of the near-full-length sequences in the three samples (human, mouse, Guerrero Negro). Each line shows one of the sequences that represents a bubble in the other panels. (b) and (c) Cluster recovery rate for the clipped sequences using all three data sets, or only the mouse data set, respectively. The size of each bubble is proportional to the recovery rate, and the number inside each bubble shows the recovery rate (i.e. the fraction of nodes in the cluster that were recovered using the clipped sequences). The _x_-axis shows the starting primer, and the _y_-axis shows the length of each amplicon. Surprisingly, although longer amplicons generally gave better cluster recoveries, some long amplicons gave very poor cluster recovery (e.g. F343-R1114 recovered only 47% of the nodes in the cluster diagram for the mouse data set). (d) and (e) Pearson correlation coefficients between the pairwise UniFrac distance scores using the full-length sequences and each set of clipped sequences from all three data sets, or from only the mouse data set, respectively. In general, the correlation between the UniFrac distances was very high even when the cluster recovery was low, suggesting that UniFrac distances are robust to primer choices (although the details of the clustering in the tree can be relatively sensitive, especially in nodes that were not jackknife-supported). Results for the Guerrero Negro data set and the human data set alone were essentially identical (data not shown).
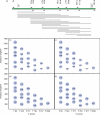
Figure 3.
UniFrac analysis of short clipped sequences simulating 454 reads, using data from all three sequence sets (human, mouse, Guerrero Negro). (a) Diagram showing clipped reads of 100, 150, 200 and 250 bases starting with each of the forward and reverse primers. Note that F1099 + 250 is not available because it exceeds the end of the near-full-length sequences we used for the analysis. (b) Correlation in UniFrac distances between jackknifed data sets and full data sets (ranging from 0, no correlation, to 1, perfect correlation). Size of bubble reflects average strength of correlation. Note that the _y_-axis on this plot ranges from 0.88 to 1, so all the correlations are very strong. The _x_-axis shows fraction of sequences retained in the jackknifing. Box plots show quartiles, medians, 95% quantiles and outliers for n = 100 jackknife replicates. (c) Cluster recovery using the same jackknifed data as (b). Note that cluster recovery is always much lower and more variable than distance recovery, indicating that many of the details of the clustering are not supported by jackknifing. (d) Cluster recovery from each primer for each read length. Best primer at each read length is shown in green; worst is shown in red. Number inside each bubble indicates the cluster recovery (size of each bubble is also proportional to cluster recovery, same scale as (b) above. (e) UniFrac PCoA clustering of the full-length sequences (legend key: hmn = human, A, B and C are three separate individuals (12); mus = mouse, M1, M2 and M3 are the three different mothers and their offspring; GN = Guerrero Negro, 10 samples are 10 different sediment layers from shallowest to deepest). (f) UniFrac PCoA clustering of an example of good cluster recovery, F517 with 200-base reads. Note that the clustering is almost identical to that of the full-length sequences, with a slight rotation of the coordinate axes, and the relative ordering of points within each cluster is preserved. (g) UniFrac PCoA clustering of an example of poor cluster recovery, R1114 with 200-base reads. The human samples are apparently split into two separate groups, suggesting the wrong biological conclusion.
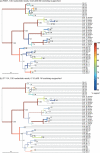
Figure 4.
UniFrac hierarchical clustering recoveries from a good and a bad primer. Data shown are from 100-base reads starting at R357 and F1114 respectively, using the cluster diagram obtained from full-length sequences as a reference. Jackknife values are shown for each node (100 replicates), and edges are shown colored by jackknife values (gray for < 60%; color bar shows scale for values above 60%). Recovered nodes are marked with an asterisk and their edges are indicated with heavy lines. R357 (a) recovers essentially all the biological signal, including the grouping of the samples from the three human individuals A, B and C, the layer structure of the Guerrero Negro microbial mat, and the clustering of mice by mother. In contrast, F1114 (b) is able only to differentiate the three general environment types from each other, and fails to recapture many nodes that are jackknife supported at the 90% level and above.
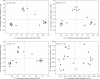
Figure 5.
UniFrac PCoA analysis of full-length human sequences, and three 100-base clipped sequence sets simulating 454 reads. The three different individuals are colored separately, with the same coloring applied to all three graphs. R357 and F917 recapture the overall pattern (discrete clusters for each individual) extremely well. In contrast, R1114 distorts the pattern substantially, and suggests separation of the three samples only along PC1. The percentage next to each primer number indicates the percentage of nodes in the hierarchical clustering on the full-length sequences that was recovered in the clipped sequences.
References
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008759/GM/NIGMS NIH HHS/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- T32GM065103/GM/NIGMS NIH HHS/United States