Rapid, transcript-specific changes in splicing in response to environmental stress - PubMed (original) (raw)
Rapid, transcript-specific changes in splicing in response to environmental stress
Jeffrey A Pleiss et al. Mol Cell. 2007.
Abstract
While the core splicing machinery is highly conserved between budding yeast and mammals, the absence of alternative splicing in Saccharomyces cerevisiae raises the fundamental question of why introns have been retained in approximately 5% of the 6000 genes. Because ribosomal protein-encoding genes (RPGs) are highly overrepresented in the set of intron-containing genes, we tested the hypothesis that splicing of these transcripts would be regulated under conditions in which translation is impaired. Using a microarray-based strategy, we find that, within minutes after the induction of amino acid starvation, the splicing of the majority of RPGs is specifically inhibited. In response to an unrelated stress, exposure to toxic levels of ethanol, splicing of a different group of transcripts is inhibited, while the splicing of a third set is actually improved. We propose that regulation of splicing, like transcription, can afford rapid and specific changes in gene expression in response to the environment.
Figures
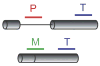
Figure 1. Splicing-Specific Microarrays
Probes targeting intron-containing genes are designed to detect the precursor species (P), the mature species (M), or the total transcript (T).
Figure 2. Regulation of Pre-mRNA Splicing in Response to Amino Acid Starvation
Time-resolved splicing profiles resulting from comparisons of (A) wild-type cells either treated with 50 mM 3AT or mock treated for 2, 4, 6, 10, 15, and 20 min; (B) prp8-1 and wild-type cells shifted from 25°C to 37°C for 5, 10, 15, 20, 25, and 30 min; and (C) wild-type cells compared to cells deleted for either RRP6, RAI1, RTT103, SKI2, UPF1, or XRN1. Also included is a comparison of two wild-type strains. The transcripts are ordered identically in all images.
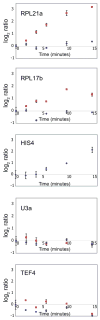
Figure 3. Quantitative RT-PCR Validates the Rapid, Transcript-Specific Downregulation of Splicing in Response to Amino Acid Starvation
The behaviors of RPL21a, RPL17b, HIS4, U3 snRNA, and TEF4 were examined using primers specific to intron regions (red squares) or exon regions (blue diamonds). Error bars are the result of triplicate measurements from a single biological sample.
Figure 4. The Splicing Response to Amino Acid Starvation Does Not Require the Activity of Gcn2
Time-resolved splicing profiles resulting from (A) wild-type cells treated with 50 mM 3AT compared to mock-treated, (B) cells deleted for GCN2 treated with 50 mM 3AT compared to mock-treated, or (C) wild-type cells treated with 50 mM 3AT compared to cells deleted for GCN2 treated with 50 mM 3AT. Included in the final set is a comparison of the wild-type strain with the _GCN2_-deleted strain in the absence of 3AT.
Figure 5. Regulation of Pre-mRNA Splicing in Response to Ethanol Toxicity
(A) Time-resolved splicing profiles resulting from comparisons of wild-type cells either treated with 10% ethanol or mock treated for 5, 10, 15, 20, 25, and 30 min. (B) Genes whose splicing is downregulated (top panel, also highlighted with a red bar in [A]) or upregulated (bottom panel, also highlighted with a green bar in [A]). (C) Behavior of genes highlighted in (B) in response to amino acid starvation.
Figure 6. Comparison of Splicing Responses to Amino Acid Starvation and Ethanol Toxicity
The order of genes is identical to that shown in Figure 2.
Comment in
- Splicing from the outside in.
Yang L, Park J, Graveley BR. Yang L, et al. Mol Cell. 2007 Sep 21;27(6):861-2. doi: 10.1016/j.molcel.2007.09.003. Mol Cell. 2007. PMID: 17889658 Free PMC article.
Similar articles
- Splicing from the outside in.
Yang L, Park J, Graveley BR. Yang L, et al. Mol Cell. 2007 Sep 21;27(6):861-2. doi: 10.1016/j.molcel.2007.09.003. Mol Cell. 2007. PMID: 17889658 Free PMC article. - Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Sapra AK, Arava Y, Khandelia P, Vijayraghavan U. Sapra AK, et al. J Biol Chem. 2004 Dec 10;279(50):52437-46. doi: 10.1074/jbc.M408815200. Epub 2004 Sep 27. J Biol Chem. 2004. PMID: 15452114 - Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays.
Clark TA, Sugnet CW, Ares M Jr. Clark TA, et al. Science. 2002 May 3;296(5569):907-10. doi: 10.1126/science.1069415. Science. 2002. PMID: 11988574 - Mechanisms and regulation of spliceosome-mediated pre-mRNA splicing in Saccharomyces cerevisiae.
Senn KA, Hoskins AA. Senn KA, et al. Wiley Interdiscip Rev RNA. 2024 Jul-Aug;15(4):e1866. doi: 10.1002/wrna.1866. Wiley Interdiscip Rev RNA. 2024. PMID: 38972853 Free PMC article. Review. - The quest for a message: budding yeast, a model organism to study the control of pre-mRNA splicing.
Meyer M, Vilardell J. Meyer M, et al. Brief Funct Genomic Proteomic. 2009 Jan;8(1):60-7. doi: 10.1093/bfgp/elp002. Epub 2009 Mar 11. Brief Funct Genomic Proteomic. 2009. PMID: 19279072 Review.
Cited by
- A quantitative, high-throughput reverse genetic screen reveals novel connections between Pre-mRNA splicing and 5' and 3' end transcript determinants.
Albulescu LO, Sabet N, Gudipati M, Stepankiw N, Bergman ZJ, Huffaker TC, Pleiss JA. Albulescu LO, et al. PLoS Genet. 2012;8(3):e1002530. doi: 10.1371/journal.pgen.1002530. Epub 2012 Mar 29. PLoS Genet. 2012. PMID: 22479188 Free PMC article. - RNA helicases in splicing.
Cordin O, Beggs JD. Cordin O, et al. RNA Biol. 2013 Jan;10(1):83-95. doi: 10.4161/rna.22547. Epub 2012 Dec 10. RNA Biol. 2013. PMID: 23229095 Free PMC article. Review. - Reciprocal hemizygosity analysis reveals that the Saccharomyces cerevisiae CGI121 gene affects lag time duration in synthetic grape must.
Li R, Deed RC. Li R, et al. G3 (Bethesda). 2021 Apr 15;11(4):jkab061. doi: 10.1093/g3journal/jkab061. G3 (Bethesda). 2021. PMID: 33681985 Free PMC article. - Analysis of Fungal Genomes Reveals Commonalities of Intron Gain or Loss and Functions in Intron-Poor Species.
Lim CS, Weinstein BN, Roy SW, Brown CM. Lim CS, et al. Mol Biol Evol. 2021 Sep 27;38(10):4166-4186. doi: 10.1093/molbev/msab094. Mol Biol Evol. 2021. PMID: 33772558 Free PMC article. - Dynamic transcriptome profiling of Bean Common Mosaic Virus (BCMV) infection in Common Bean (Phaseolus vulgaris L.).
Martin K, Singh J, Hill JH, Whitham SA, Cannon SB. Martin K, et al. BMC Genomics. 2016 Aug 11;17(1):613. doi: 10.1186/s12864-016-2976-8. BMC Genomics. 2016. PMID: 27515794 Free PMC article.
References
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. - PubMed
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. - PubMed
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
- Chen JC, Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr Genet. 2006;49:281–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases