Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking - PubMed (original) (raw)
Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking
Hayley J Newton et al. Infect Immun. 2007 Dec.
Abstract
The environmental pathogen Legionella pneumophila possesses five proteins with Sel1 repeats (SLRs) from the tetratricopeptide repeat protein family. Three of these proteins, LpnE, EnhC, and LidL, have been implicated in the ability of L. pneumophila to efficiently establish infection and/or manipulate host cell trafficking events. Previously, we showed that LpnE is important for L. pneumophila entry into macrophages and epithelial cells. In further virulence studies here, we show that LpnE is also required for efficient infection of Acanthamoeba castellanii by L. pneumophila and for replication of L. pneumophila in the lungs of A/J mice. In addition, we found that the role of LpnE in host cell invasion is dependent on the eight SLR regions of the protein. A truncated form of LpnE lacking the two C-terminal SLR domains was unable to complement the invasion defect of an lpnE mutant of L. pneumophila 130b in both the A549 and THP-1 cell lines. The lpnE mutant displayed impaired avoidance of LAMP-1 association, suggesting that LpnE influenced trafficking of the L. pneumophila vacuole, similar to the case for EnhC and LidL. We also found that LpnE was present in L. pneumophila culture supernatants and that its export was independent of both the Lsp type II secretion system and the Dot/Icm type IV secretion system. The fact that LpnE was exported suggested that the protein may interact with a eukaryotic protein. Using LpnE as bait, we screened a HeLa cell cDNA library for interacting partners, using the yeast two-hybrid system. Examination of the protein-protein interaction between LpnE and a eukaryotic protein, obscurin-like protein 1, suggested that LpnE can interact with eukaryotic proteins containing immunoglobulin-like folds via the SLR regions. This investigation has further characterized the contribution of LpnE to L. pneumophila virulence and, more specifically, the importance of the SLR regions to LpnE function.
Figures
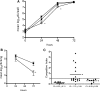
FIG. 1.
(A) L. pneumophila infection of A. castellanii. Amoebae were infected at an MOI of 0.01 with wild-type L. pneumophila 130b (▪) or the lpnE::km (○) or lpnE::km/(pMIP:lpnE) (▴) mutant. Bacterial CFU were determined at 24, 48, and 72 h postinfection and are represented as mean log10 CFU ± standard deviations for three independent experiments. *, significantly different from 130b (P = 0.034 at 24 h and P = 0.003 at 48 h [unpaired, two-tailed t test]). (B) Lung colonization of A/J mice by L. pneumophila 130b (▪) and an lpnE::km mutant (○). Strain 130b and the lpnE::km mutant were introduced into the lungs of A/J mice via intratracheal inoculation at a ratio of 1:1. Twenty-four and 72 h following infection, the numbers of CFU for 130b and the lpnE::km mutant were determined. Data are expressed as means ± standard deviations of the log10 CFU recovered per lung (n = 7 or 8). *, significantly different from 130b at 72 h (P = 0.018; unpaired, two-tailed t test). (C) CIs of derivatives of L. pneumophila 130b in mixed infections with the wild-type parent strain. The pairs tested were as follows: lpnE::km mutant versus 130b (○), lpnE::km/(pMIP:lpnE) mutant versus 130b/(pMIP) (▴), and lpnE::km/(pMIP:_lpnE_52-375) mutant versus 130b/(pMIP) (•). *, CI was significantly different from that for the lpnE::km mutant versus 130b (P < 0.05; unpaired, two-tailed t test).
FIG. 2.
(A) Immunofluorescence of LAMP-1 in A549 cells infected for 5 h with wild-type L. pneumophila 130b or the dotA::cm, lpnE::km, or lpnE::km/(pMIP:lpnE) mutant. LAMP-1 was detected with an anti-LAMP-1 mouse monoclonal antibody diluted 1/100 followed by the secondary antibody, anti-mouse-Alexa Fluor 488, diluted 1/200. Bacteria were detected with anti-lipopolysaccharide (anti-LPS) antibodies raised in rabbits and diluted 1/50, followed by the secondary antibody Alexa Fluor 594 diluted 1/200. (B) Immunofluorescence of LAMP-1 in A549 cells infected for 24 h with wild-type L. pneumophila 130b or the dotA::cm, lpnE::km, or lpnE::km/(pMIP:lpnE) mutant. Primary and secondary antibodies were the same as for panel A.
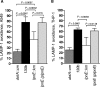
FIG. 3.
Percentage of _Legionella_-containing vacuoles that avoided LAMP-1 after infection of A549 cells (A) and THP-1 cells (B) for 5 h. P values of <0.05 (unpaired two-tailed t test) are indicated. LAMP-1 avoidance was scored blind according to the staining patterns indicated in Fig. 4A.
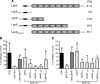
FIG. 4.
(A) Schematic representation of L. pneumophila SLR protein LpnE and truncated variants created for this study. Shaded rectangles represent the SLR regions, and black rectangles signify the predicted N-terminal 22-amino-acid signal peptide. The truncations were used to complement L. pneumophila lpnE::km, and resulting strains were examined for uptake by THP-1 macrophages (B) and A549 epithelial cells (C). Data are expressed as percentages of the amount of inoculum that was intracellular following a 2-h infection and 1-h gentamicin treatment and are means ± standard deviations for at least three independent experiments. *, significantly different from the lpnE::km mutant (P < 0.05; unpaired two-tailed t test).
FIG. 5.
Immunoblot analysis of culture supernatants precipitated with TCA and detected with anti-LpnE, anti-Lpg1905, and anti-RpoB antibodies. (A) Stationary-phase TCA-precipitated culture supernatants and whole-cell lysate samples from derivatives of L. pneumophila 130b were separated by SDS-polyacrylamide gel electrophoresis. Lane 1, L. pneumophila 130b; lane 2, lpnE::km mutant; lane 3, lpnE::km/(pMIP:lpnE) mutant; lane 4, Δ_dotA_ mutant; lane 5, Δ_lspDE_ mutant. (B) TCA-precipitated stationary-phase culture supernatants. Lane 1, L. pneumophila lpnE::km/(pMIP:lpnE) mutant; lane 2, lpnE::km/(pMIP:_lpnE_1-51) mutant; lane 3, lpnE::km/(pMIP:_lpnE_1-122) mutant; lane 4, lpnE::km/(pMIP:_lpnE_1-266) mutant; lane 5, lpnE::km/(pMIP:_lpnE_52-375) mutant. The presence of both LpnE1-266 and LpnE52-375 in the culture supernatant is indicated by arrows and the predicted molecular size of each protein.
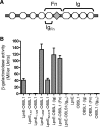
FIG. 6.
(A) Domain organization of OBSL1, comprising several Ig-like domains (circles) and one Fn domain (diamond). The regions of OBSL1 subcloned for protein-protein interaction studies, namely, Ig, Fn, and IgFn, are indicated. (B) Protein-protein interactions were assessed using the yeast two-hybrid system β-galactosidase reporter assay. Full-length LpnE, LpnE1-266, and LpnE52-375 showed strong interactions with OBSL1, as did full-length LpnE with two of the truncated forms of OBSL1 but not with IgFn.
Similar articles
- Implication of proteins containing tetratricopeptide repeats in conditional virulence phenotypes of Legionella pneumophila.
Bandyopadhyay P, Sumer EU, Jayakumar D, Liu S, Xiao H, Steinman HM. Bandyopadhyay P, et al. J Bacteriol. 2012 Jul;194(14):3579-88. doi: 10.1128/JB.00399-12. Epub 2012 May 4. J Bacteriol. 2012. PMID: 22563053 Free PMC article. - The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein.
Voth KA, Chung IYW, van Straaten K, Li L, Boniecki MT, Cygler M. Voth KA, et al. FEBS J. 2019 Feb;286(4):710-725. doi: 10.1111/febs.14710. Epub 2018 Dec 20. FEBS J. 2019. PMID: 30479037 - The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth.
Vincent CD, Vogel JP. Vincent CD, et al. Mol Microbiol. 2006 Aug;61(3):596-613. doi: 10.1111/j.1365-2958.2006.05243.x. Epub 2006 Jun 27. Mol Microbiol. 2006. PMID: 16803597 - Sel1-like repeat proteins in signal transduction.
Mittl PR, Schneider-Brachert W. Mittl PR, et al. Cell Signal. 2007 Jan;19(1):20-31. doi: 10.1016/j.cellsig.2006.05.034. Epub 2006 Jul 25. Cell Signal. 2007. PMID: 16870393 Review. - Effector proteins translocated by Legionella pneumophila: strength in numbers.
Ninio S, Roy CR. Ninio S, et al. Trends Microbiol. 2007 Aug;15(8):372-80. doi: 10.1016/j.tim.2007.06.006. Epub 2007 Jul 13. Trends Microbiol. 2007. PMID: 17632005 Review.
Cited by
- Structure-Function Analysis of DipA, a Francisella tularensis Virulence Factor Required for Intracellular Replication.
Chong A, Child R, Wehrly TD, Rockx-Brouwer D, Qin A, Mann BJ, Celli J. Chong A, et al. PLoS One. 2013 Jun 26;8(6):e67965. doi: 10.1371/journal.pone.0067965. Print 2013. PLoS One. 2013. PMID: 23840797 Free PMC article. - Functional characterization of two secreted SEL1L isoforms capable of exporting unassembled substrate.
Cattaneo M, Lotti LV, Martino S, Cardano M, Orlandi R, Mariani-Costantini R, Biunno I. Cattaneo M, et al. J Biol Chem. 2009 Apr 24;284(17):11405-15. doi: 10.1074/jbc.M805408200. Epub 2009 Feb 9. J Biol Chem. 2009. PMID: 19204006 Free PMC article. - Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes?
Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Catalán-Nájera JC, et al. Virulence. 2017 Oct 3;8(7):1111-1123. doi: 10.1080/21505594.2017.1317412. Epub 2017 Apr 12. Virulence. 2017. PMID: 28402698 Free PMC article. Review. - Cell biology of infection by Legionella pneumophila.
Xu L, Luo ZQ. Xu L, et al. Microbes Infect. 2013 Feb;15(2):157-67. doi: 10.1016/j.micinf.2012.11.001. Epub 2012 Nov 14. Microbes Infect. 2013. PMID: 23159466 Free PMC article. Review. - Tol-Pal System and Rgs Proteins Interact to Promote Unipolar Growth and Cell Division in Sinorhizobium meliloti.
Krol E, Yau HCL, Lechner M, Schäper S, Bange G, Vollmer W, Becker A. Krol E, et al. mBio. 2020 Jun 30;11(3):e00306-20. doi: 10.1128/mBio.00306-20. mBio. 2020. PMID: 32605980 Free PMC article.
References
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- Bartel, P. L., C.-T. Chien, R. Sternglanz, and S. R. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions, p. 153-179. In D. A. Hartley (ed.), Cellular interactions in development: a cellular approach. Oxford University Press, Oxford, United Kingdom.
- Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous