DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes - PubMed (original) (raw)
DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes
Karsten Rippe et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Chromatin-remodeling complexes can translocate nucleosomes along the DNA in an ATP-coupled reaction. This process is an important regulator of all DNA-dependent processes because it determines whether certain DNA sequences are found in regions between nucleosomes with increased accessibility for other factors or wrapped around the histone octamer complex. In a comparison of seven different chromatin-remodeling machines (ACF, ISWI, Snf2H, Chd1, Mi-2, Brg1, and NURF), it is demonstrated that these complexes can read out DNA sequence features to establish specific nucleosome-positioning patterns. For one of the remodelers, ACF, we identified a 40-bp DNA sequence element that directs nucleosome positioning. Furthermore, we show that nucleosome positioning by the remodelers ACF and Chd1 is determined by a reduced affinity to the end product of the translocation reaction. The results suggest that the linkage of differential remodeling activities with the intrinsic binding preferences of nucleosomes can result in establishing distinct chromatin structures that depend on the DNA sequence and define the DNA accessibility for other protein factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Mammalian chromatin-remodeling complexes are highly diverse. (Left) The Snf2 family members present in humans, with the number of individual proteins within a subfamily in parentheses. The 11 subfamilies marked in red were shown to possess ATP-dependent chromatin-remodeling activities. Each of these subfamilies comprises many different members. (Right) The multiple ISWI ATPases complexes known to date. There are four ISWI subgroups (green boxes), and the known chromatin-remodeling complexes containing these ATPases are listed. Complexes in light gray type were shown to exist with the alternative ISWI-type ATPases.
Fig. 2.
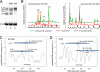
Chromatin-remodeling complexes position nucleosomes in a DNA sequence-specific manner. The nucleosome substrate was reconstituted by salt dialysis on a radioactively labeled 350-bp fragment carrying the hsp70 promoter. A mixture of a single nucleosome at five different major positions, indicated as N1, N2, N3, N4, and N4′, was obtained (28). This mixed nucleosome population (lane 1) was used as the same substrate for all seven remodelers shown. The endpoint of the nucleosome translocation reaction obtained after incubation for 90 min at 26°C in the presence of ATP is shown for recombinant Brg1 (lane 2), Chd1 (lane 3), ISWI (lane 4), Snf2H (lane 5), Mi-2 (lane 6), ACF (lane 7), and NURF (lane 8).
Fig. 3.
A short DNA element can direct ACF-dependent nucleosome positioning. (A) Remodeling reaction with ACF or ISWI with a nucleosome substrate containing a 253-bp-long DNA fragment (K3 DNA) from the pT-K3 plasmid. After nucleosome assembly by salt dialysis on the K3 DNA, a mixed population of a single nucleosome with three main positions (N1, N2, and N4) and one minor position (N3, lane 1) was obtained. This substrate was used in a remodeling reaction with ISWI (lane 2) or ACF (lane 3). (B) High-resolution mapping of the remodeler-dependent nucleosome positions on the K3 DNA substrate. MNase protection and subsequent primer extension reactions were conducted. Scans for the primer extension reactions (Left, forward primer; Right, reverse primer) are shown for the nucleosomal input substrate (green, corresponding to A, lane 1) and the remodeling reaction for ACF (red, corresponding to A, lane 3). The black curve shows a 10-bp DNA marker. The same analysis was conducted with ISWI (data not shown). The peaks reflect nucleosomes positioned adjacent to this site. Considering that 147 bp of DNA are protected by the nucleosome, the major nucleosome positions were identified as 37/45 to 187/195 for N1, 25 to 175 for N2, and 0/7 to 151/157 for N4. (C) The ACF- and ISWI-dependent nucleosome positions determined on the 253-bp K3 DNA fragment were plotted together with the predicted DNA curvature. The black arrow refers to the 40-bp DNA sequence encompassing the region of maximal DNA curvature from the rDNA sequence that was cloned into the K3 DNA. (D) Same analysis as in C, but for the 248-bp rDNA promoter fragment with the previously determined nucleosome sites (30, 31).
Fig. 4.
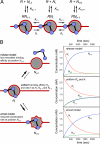
Mechanisms of nucleosome positioning by chromatin-remodeling complexes. (A) Scheme for the remodeling reaction. Three positions on the DNA (i − 1, i, and i + 1) are considered. The remodeler R can bind to a nucleosome N at each of these three positions with a dissociation constant _K_d. Translocation to or from these positions occurs with rate constants k as described in the text. (B) Two models for nucleosome positioning are depicted. (Right) Corresponding time course of the concentrations of nucleosomes. The reaction starts with all nucleosomes at position i at an initial concentration of 2.5 × 10−9 M. If all binding constants and translocation rate constants are identical (uniform _K_d and k), a homogenous distribution is obtained in equilibrium, where one-third of the nucleosomes is at positions i − 1, i, and i + 1, as expected. (Left) In the release model, the binding affinity to the nucleosome at position i + 1 is reduced by a 10-fold higher value of _K_d,i+1, compared with positions i and i − 1. This result leads to a distribution in which ≈80% of the nucleosomes are at this site when an equilibrium is reached. For the arrest model, the rate constant _k_−1 that translocates the nucleosome from position i + 1 back to position i is 10 times reduced compared with the other translocation rates. Again ≈80% of the nucleosomes will be positioned at site i + 1 as the reaction reaches a steady state. The kinetic simulations were conducted with concentrations similar to those used in the in vitro reaction of ≈1 remodeler per 50 nucleosomes.
Fig. 5.
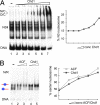
ACF and Chd1 position nucleosomes according to the release model. Nucleosome position-dependent differences in the affinity of the remodeling complexes to the nucleosomal substrate were analyzed by EMSAs. (A) (Left) A mixed nucleosomal species reconstituted on the hsp70 DNA (lane 1) was incubated with increasing concentrations of Chd1 (lanes 2–7) in the absence of ATP. The position of the appearing DNA–Chd1 (D/C) and the nucleosome–Chd1 (N/C) complexes are indicated. The position of the N3 nucleosome is shown by a black box. This position also is the preferred endpoint of the remodeling reaction (see Fig. 2). (Right) Percentage of nucleosomes at position N3 (radioactivity in the N3 band divided through the sum of the radioactivity of all nucleosome bands) is plotted versus the Chd1 concentration. An increase of the N3 fraction is apparent, suggesting that this site is the lowest affinity binding site for Chd1 with this substrate. (B) Chd1 and ACF binding to nucleosomes reconstituted at the rDNA promoter fragment. A purified mixture of nucleosomes positioned at the center and the border of the rDNA fragment (lane 1) was incubated with increasing concentrations of ACF (lanes 2 and 3) or Chd1 (lanes 4 and 5). The position of remodeler–nucleosome complexes (N/R) is indicated. The graph represents the fraction of nucleosomes at the center position with increasing concentrations of Chd1 or ACF. This lower affinity binding site also is the preferred endpoint of the reaction as shown in
SI Fig. 7_B_
.
Similar articles
- Control of nucleosome positions by DNA sequence and remodeling machines.
Schnitzler GR. Schnitzler GR. Cell Biochem Biophys. 2008;51(2-3):67-80. doi: 10.1007/s12013-008-9015-6. Epub 2008 Jun 10. Cell Biochem Biophys. 2008. PMID: 18543113 Review. - Antagonistic forces that position nucleosomes in vivo.
Whitehouse I, Tsukiyama T. Whitehouse I, et al. Nat Struct Mol Biol. 2006 Jul;13(7):633-40. doi: 10.1038/nsmb1111. Epub 2006 Jul 2. Nat Struct Mol Biol. 2006. PMID: 16819518 - Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences.
Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Thåström A, et al. J Mol Biol. 1999 Apr 30;288(2):213-29. doi: 10.1006/jmbi.1999.2686. J Mol Biol. 1999. PMID: 10329138 - New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning.
Lowary PT, Widom J. Lowary PT, et al. J Mol Biol. 1998 Feb 13;276(1):19-42. doi: 10.1006/jmbi.1997.1494. J Mol Biol. 1998. PMID: 9514715 - DNA recognition and nucleosome organization.
Travers A, Drew H. Travers A, et al. Biopolymers. 1997;44(4):423-33. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. Biopolymers. 1997. PMID: 9782778 Review.
Cited by
- Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner.
Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP. Moshkin YM, et al. Mol Cell Biol. 2012 Feb;32(3):675-88. doi: 10.1128/MCB.06365-11. Epub 2011 Nov 28. Mol Cell Biol. 2012. PMID: 22124157 Free PMC article. - PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor.
Ho KK, Zhang H, Golden BL, Ogas J. Ho KK, et al. Biochim Biophys Acta. 2013 Feb;1829(2):199-210. doi: 10.1016/j.bbagrm.2012.10.011. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128324 Free PMC article. - Dynamic and selective nucleosome repositioning during endotoxin tolerance.
El Gazzar M, Liu T, Yoza BK, McCall CE. El Gazzar M, et al. J Biol Chem. 2010 Jan 8;285(2):1259-71. doi: 10.1074/jbc.M109.067330. Epub 2009 Nov 9. J Biol Chem. 2010. PMID: 19901031 Free PMC article. - Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities.
Teif VB, Rippe K. Teif VB, et al. Nucleic Acids Res. 2009 Sep;37(17):5641-55. doi: 10.1093/nar/gkp610. Epub 2009 Jul 22. Nucleic Acids Res. 2009. PMID: 19625488 Free PMC article. - Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions.
Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Morris SA, et al. Nat Struct Mol Biol. 2014 Jan;21(1):73-81. doi: 10.1038/nsmb.2718. Epub 2013 Dec 8. Nat Struct Mol Biol. 2014. PMID: 24317492 Free PMC article.
References
- Muchardt C, Yaniv M. J Mol Biol. 1999;293:187–198. - PubMed
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Nature. 1999;402:418–421. - PubMed
- Becker PB, Hörz W. Annu Rev Biochem. 2002;71:247–273. - PubMed
- Roth SY, Shimizu M, Johnson L, Grunstein M, Simpson RT. Genes Dev. 1992;6:411–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous