Genome-wide expression and location analyses of the Candida albicans Tac1p regulon - PubMed (original) (raw)
Genome-wide expression and location analyses of the Candida albicans Tac1p regulon
Teresa T Liu et al. Eukaryot Cell. 2007 Nov.
Abstract
A major mechanism of azole resistance in Candida albicans is overexpression of the genes encoding the ATP binding cassette transporters Cdr1p and Cdr2p due to gain-of-function mutations in Tac1p, a transcription factor of the zinc cluster family. To identify the Tac1p regulon, we analyzed four matched sets of clinical isolates representing the development of CDR1- and CDR2-mediated azole resistance by using gene expression profiling. We identified 31 genes that were consistently up-regulated with CDR1 and CDR2, including TAC1 itself, and 12 consistently down-regulated genes. When a resistant strain deleted for TAC1 was examined similarly, expression of almost all of these genes returned to levels similar to those in the matched azole-susceptible isolate. Using genome-wide location (ChIP-chip) analysis (a procedure combining chromatin immunoprecipitation with hybridization to DNA intergenic microarrays), we found 37 genes whose promoters were bound by Tac1p in vivo, including CDR1 and CDR2. Sequence analysis identified nine new genes whose promoters contain the previously reported Tac1p drug-responsive element (CGGN(4)CGG), including TAC1. In total, there were eight genes whose expression was modulated in the four azole-resistant clinical isolates in a TAC1-dependent manner and whose promoters were bound by Tac1p, qualifying them as direct Tac1p targets: CDR1, CDR2, GPX1 (putative glutathione peroxidase), LCB4 (putative sphingosine kinase), RTA3 (putative phospholipid flippase), and orf19.1887 (putative lipase), as well as IFU5 and orf19.4898 of unknown function. Our results show that Tac1p binds under nonactivating conditions to the promoters of its targets, including to its own promoter. They also suggest roles for Tac1p in regulating lipid metabolism (mobilization and trafficking) and oxidative stress response in C. albicans.
Figures
FIG. 1.
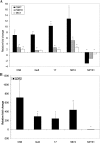
Quantitative real-time RT-PCR analysis of selected genes differentially expressed in the microarray experiments. (A) Genes differentially expressed in each of the azole-resistant clinical isolates compared to their parent isolates. (B) CDR2 gene expression in each of the azole-resistant clinical isolates. Asterisks denote statistical significance by the t test (P ≤ 0.05). Error bars denote standard deviations.
FIG. 2.
Chromosomal tagging of Tac1p. (A) PCR epitope-tagging strategy for C-terminally tagging transcription factor Tac1p with the triple HA epitope. (Top) Primers (100 nucleotides) (see Materials and Methods) were designed such that the 5′ 84 bases of the forward (FWD) and reverse (REV) primers are homologous to sequences of the TAC1 gene and the 3′ 16 bases are complementary and in-frame to unique sequences (open boxes) in the tagging cassette which contains the C. albicans URA3 marker (Ca_URA3_, light-gray box) flanked by direct repeats of the HA3-encoding sequences (HA, black boxes). The Tac1p stop codon is indicated by the asterisk. (Middle) PCR amplification results in a fragment whose ends include the primer sequences, allowing integration by homologous recombination of the tagging cassette upstream of the TAC1 3′-untranslated region (thick horizontal line). The orientation of the TAC1 ORF (dark-gray box) is indicated by the arrow. (Bottom) C. albicans URA3 marker excision results in the final product, _TAC1_-HA3. (B) Schematic representations of the _TAC1_-1 (orf19.3188) and _TAC1_-2 (orf19.10700) alleles (gray boxes) in strain CAI4. Sizes of the HindIII (HIII)/EcoRV (RV) double-digested fragments detected by the TAC1 probe (top, thick line) used for the Southern blot experiment are given for the _TAC1_-2 allele following integration of the HA3-tagging cassette (open box) and excision of the C. albicans URA3 marker through HA3 recombination. (C) Southern blot analysis of genomic DNA from the CAI4 strain and its URA3 preexcision (SZY51) and postexcision (SZY63) derivatives, digested with EcoRV and HindIII and hybridized with the TAC1 probe shown in panel B. Marker sizes are indicated on the left. (D) Western blot analysis of protein extracts from strains CAI4 and the Tac1p-HA3 integrant SZY63 with an anti-HA monoclonal antibody. Molecular size markers are indicated on the left.
FIG. 3.
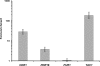
In vivo enrichment of Tac1p binding at the CDR1, PDR16, and TAC1 promoters, determined using Q-PCR. The CAI4 and SZY63 strains were submitted to ChIP (three biological replicates), and the recovered DNA samples were analyzed by Q-PCR using Universal ProbeLibrary probes (Roche) for the PDR16, TAC1, SPS4, and FUR1 promoters or a TaqMan probe (IDT) for the CDR1 promoter. Enrichments (_n_-fold) are presented in log scale: 3.8 for the PDR16 promoter (95% confidence interval of 4.0, 5.0), 28.8 for the CDR1 promoter (95% confidence interval of 21.4, 38.9), 189.3 for the TAC1 promoter (95% confidence interval of 128.6, 278.6), and 1.1 for the FUR1 promoter (95% confidence interval of 0.9, 1.3), which was used as a negative control. Error bars denote standard deviations.
Similar articles
- TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2.
Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. Coste AT, et al. Eukaryot Cell. 2004 Dec;3(6):1639-52. doi: 10.1128/EC.3.6.1639-1652.2004. Eukaryot Cell. 2004. PMID: 15590837 Free PMC article. - Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system.
Coste AT, Crittin J, Bauser C, Rohde B, Sanglard D. Coste AT, et al. Eukaryot Cell. 2009 Aug;8(8):1250-67. doi: 10.1128/EC.00069-09. Epub 2009 Jun 26. Eukaryot Cell. 2009. PMID: 19561319 Free PMC article. - The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans.
Znaidi S, De Deken X, Weber S, Rigby T, Nantel A, Raymond M. Znaidi S, et al. Mol Microbiol. 2007 Oct;66(2):440-52. doi: 10.1111/j.1365-2958.2007.05931.x. Mol Microbiol. 2007. PMID: 17897373 - The H741D mutation in Tac1p contributes to the upregulation of CDR1 and CDR2 expression in Candida albicans.
Liu JY, Wei B, Wang Y, Shi C, Li WJ, Zhao Y, Meng LN, Xiang MJ. Liu JY, et al. Braz J Microbiol. 2020 Dec;51(4):1553-1561. doi: 10.1007/s42770-020-00336-8. Epub 2020 Jul 9. Braz J Microbiol. 2020. PMID: 32648240 Free PMC article. - The TAC1 Gene in Candida albicans: Structure, Function, and Role in Azole Resistance: A Mini-Review.
Mahdizade AH, Hoseinnejad A, Ghazanfari M, Boozhmehrani MJ, Bahreiny SS, Abastabar M, Galbo R, Giuffrè L, Haghani I, Romeo O. Mahdizade AH, et al. Microb Drug Resist. 2024 Jul;30(7):288-296. doi: 10.1089/mdr.2023.0334. Epub 2024 May 21. Microb Drug Resist. 2024. PMID: 38770776 Review.
Cited by
- A Double-Edged Sword: Aneuploidy is a Prevalent Strategy in Fungal Adaptation.
Tsai HJ, Nelliat A. Tsai HJ, et al. Genes (Basel). 2019 Oct 10;10(10):787. doi: 10.3390/genes10100787. Genes (Basel). 2019. PMID: 31658789 Free PMC article. Review. - Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance.
Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, Raymond M. Znaidi S, et al. Eukaryot Cell. 2008 May;7(5):836-47. doi: 10.1128/EC.00070-08. Epub 2008 Apr 4. Eukaryot Cell. 2008. PMID: 18390649 Free PMC article. - A Hyperactive Form of the Zinc Cluster Transcription Factor Stb5 Causes YOR1 Overexpression and Beauvericin Resistance in Candida albicans.
Ramírez-Zavala B, Manz H, Englert F, Rogers PD, Morschhäuser J. Ramírez-Zavala B, et al. Antimicrob Agents Chemother. 2018 Nov 26;62(12):e01655-18. doi: 10.1128/AAC.01655-18. Print 2018 Dec. Antimicrob Agents Chemother. 2018. PMID: 30249688 Free PMC article. - A Proteomic Landscape of Candida albicans in the Stepwise Evolution to Fluconazole Resistance.
Song N, Zhou X, Li D, Li X, Liu W. Song N, et al. Antimicrob Agents Chemother. 2022 Apr 19;66(4):e0210521. doi: 10.1128/aac.02105-21. Epub 2022 Mar 28. Antimicrob Agents Chemother. 2022. PMID: 35343782 Free PMC article. - Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans.
Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, Nantel A. Sellam A, et al. Mol Biol Cell. 2009 May;20(9):2389-400. doi: 10.1091/mbc.e08-11-1093. Epub 2009 Mar 11. Mol Biol Cell. 2009. PMID: 19279142 Free PMC article.
References
- Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. - PubMed
- Avery, A. M., and S. V. Avery. 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276:33730-33735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous