Capturing heterogeneity in gene expression studies by surrogate variable analysis - PubMed (original) (raw)
Capturing heterogeneity in gene expression studies by surrogate variable analysis
Jeffrey T Leek et al. PLoS Genet. 2007 Sep.
Abstract
It has unambiguously been shown that genetic, environmental, demographic, and technical factors may have substantial effects on gene expression levels. In addition to the measured variable(s) of interest, there will tend to be sources of signal due to factors that are unknown, unmeasured, or too complicated to capture through simple models. We show that failing to incorporate these sources of heterogeneity into an analysis can have widespread and detrimental effects on the study. Not only can this reduce power or induce unwanted dependence across genes, but it can also introduce sources of spurious signal to many genes. This phenomenon is true even for well-designed, randomized studies. We introduce "surrogate variable analysis" (SVA) to overcome the problems caused by heterogeneity in expression studies. SVA can be applied in conjunction with standard analysis techniques to accurately capture the relationship between expression and any modeled variables of interest. We apply SVA to disease class, time course, and genetics of gene expression studies. We show that SVA increases the biological accuracy and reproducibility of analyses in genome-wide expression studies.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Impact of Expression Heterogeneity
One thousand gene expression datasets containing EH were simulated, tested, and ranked for differential expression as detailed in Simulated Examples. (A) A boxplot of the standard deviation of the ranks of each gene for differential expression over repeated simulated studies. Results are shown for analyses that ignore expression heterogeneity (Unadjusted), take expression heterogeneity into account by SVA (Adjusted), and for simulated data unaffected by expression heterogeneity (Ideal). (B) For each simulated dataset, a Kolmogorov-Smirnov test was employed to assess whether the _p-_values of null genes followed the correct null Uniform distribution (Text S1). A quantile–quantile plot of the 1,000 Kolmogorov-Smirnov _p-_values are shown for the SVA-adjusted analysis (solid line) and the unadjusted analysis (dashed line). It can be seen that the SVA-adjusted analysis provides correctly distributed null _p-_values, whereas the unadjusted analysis does not due to EH. (C) A plot of expected true positives versus FDR for the SVA-adjusted (solid) and -unadjusted (dashed) analyses. The SVA-adjusted analysis shows increased power to detect true differential expression.
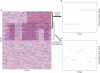
Figure 2. Example of Expression Heterogeneity
(A) A heatmap of a simulated microarray study consisting of 1,000 genes measured on 20 arrays. (B) Genes 1–300 in this simulated study are differentially expressed between two hypothetical treatment groups; here the two groups are shown as an indicator variable for each array. (C) Genes 201–500 in each simulated study are affected by an independent factor that causes EH. This factor is distinct from, but possibly correlated with, the group variable. Here, the factor is shown as a quantitative variable, but it could also be an indicator variable or some linear or nonlinear function of the covariates.
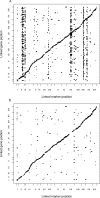
Figure 3. SVA Captures EH Due to Genotype
(A) A plot of significant linkage peaks (_p_-value < 1e−7) for expression QTL in the Brem et al. [10,21] study by marker location (_x_-axis) and expression trait location (_y_-axis). (B) Significant linkage peaks (_p_-value < 1e−7) after adjusting for surrogate variables. Large _trans_-linkage peaks on Chromosomes II, III, VII, XII, XIV, and XV have been eliminated without reducing _cis_-linkage peaks.
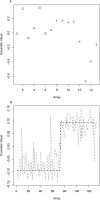
Figure 4. Surrogate Variables from Human Studies
(A) A plot of the top surrogate variable estimated from the breast cancer data [22]. The BRCA1 group is relatively homogeneous (triangles), but the BRCA2 group shows substantial heterogeneity (pluses). (B) A plot of tissue type versus array for the Rodwell et al. [7] study (dotted line) and the top surrogate variable estimated from the expression data when tissue was ignored (dashed line). There is strong correlation between the top surrogate variable and the tissue type variable.
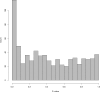
Figure 5. Null _p_-Values under Heterogeneity
A histogram of the null _p-_values from a single simulated experiment affected by heterogeneity. The distribution of these _p-_values appears identical to a complete set of _p-_values from an experiment that is not subject to heterogeneity. Therefore, it is not possible to identify and account for heterogeneity by analyzing one-dimensional _p-_values or test-statistics (see also Text S1).
Similar articles
- SVAw - a web-based application tool for automated surrogate variable analysis of gene expression studies.
Pirooznia M, Seifuddin F, Goes FS, Leek JT, Zandi PP. Pirooznia M, et al. Source Code Biol Med. 2013 Mar 11;8(1):8. doi: 10.1186/1751-0473-8-8. Source Code Biol Med. 2013. PMID: 23497726 Free PMC article. - Surrogate variable analysis using partial least squares (SVA-PLS) in gene expression studies.
Chakraborty S, Datta S, Datta S. Chakraborty S, et al. Bioinformatics. 2012 Mar 15;28(6):799-806. doi: 10.1093/bioinformatics/bts022. Epub 2012 Jan 11. Bioinformatics. 2012. PMID: 22238271 - Use of expression data and the CGEMS genome-wide breast cancer association study to identify genes that may modify risk in BRCA1/2 mutation carriers.
Walker LC, Waddell N, Ten Haaf A; kConFab Investigators; Grimmond S, Spurdle AB. Walker LC, et al. Breast Cancer Res Treat. 2008 Nov;112(2):229-36. doi: 10.1007/s10549-007-9848-5. Epub 2007 Dec 20. Breast Cancer Res Treat. 2008. PMID: 18095154 - Gene analysis techniques and susceptibility gene discovery in non-BRCA1/BRCA2 familial breast cancer.
Aloraifi F, Boland MR, Green AJ, Geraghty JG. Aloraifi F, et al. Surg Oncol. 2015 Jun;24(2):100-9. doi: 10.1016/j.suronc.2015.04.003. Epub 2015 Apr 13. Surg Oncol. 2015. PMID: 25936246 Review. - Histopathology of BRCA1- and BRCA2-associated breast cancer.
Honrado E, Benítez J, Palacios J. Honrado E, et al. Crit Rev Oncol Hematol. 2006 Jul;59(1):27-39. doi: 10.1016/j.critrevonc.2006.01.006. Epub 2006 Mar 10. Crit Rev Oncol Hematol. 2006. PMID: 16530420 Review.
Cited by
- MethylCallR : a comprehensive analysis framework for Illumina Methylation Beadchip.
Yang HH, Han MR. Yang HH, et al. Sci Rep. 2024 Nov 7;14(1):27026. doi: 10.1038/s41598-024-77914-5. Sci Rep. 2024. PMID: 39506033 - Sensitivity to Unobserved Confounding in Studies with Factor-structured Outcomes.
Zheng J, Wu J, D'Amour A, Franks A. Zheng J, et al. J Am Stat Assoc. 2024;119(547):2026-2037. doi: 10.1080/01621459.2023.2240053. Epub 2023 Sep 25. J Am Stat Assoc. 2024. PMID: 39493289 - Correction of Batch Effect in Gut Microbiota Profiling of ASD Cohorts from Different Geographical Origins.
Scanu M, Del Chierico F, Marsiglia R, Toto F, Guerrera S, Valeri G, Vicari S, Putignani L. Scanu M, et al. Biomedicines. 2024 Oct 15;12(10):2350. doi: 10.3390/biomedicines12102350. Biomedicines. 2024. PMID: 39457661 Free PMC article. - Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage.
Vignolle GA, Bauerstätter P, Schönthaler S, Nöhammer C, Olischar M, Berger A, Kasprian G, Langs G, Vierlinger K, Goeral K. Vignolle GA, et al. Int J Mol Sci. 2024 Sep 25;25(19):10304. doi: 10.3390/ijms251910304. Int J Mol Sci. 2024. PMID: 39408633 Free PMC article. - Thinking points for effective batch correction on biomedical data.
Hui HWH, Kong W, Goh WWB. Hui HWH, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae515. doi: 10.1093/bib/bbae515. Brief Bioinform. 2024. PMID: 39397427 Free PMC article.
References
- Klebanov L, Yakovlev A. Treating expression levels of different genes as a sample in microarray data analysis: is it worth a risk? Stat Appl Genet Mol Biol. 2006;5:art9. - PubMed
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000;7:819–837. - PubMed
- Kerr MK, Churchill GA. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases