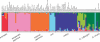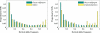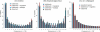Genome-wide patterns of nucleotide polymorphism in domesticated rice - PubMed (original) (raw)
doi: 10.1371/journal.pgen.0030163. Epub 2007 Aug 6.
Scott H Williamson, Ryan D Hernandez, Adam Boyko, Adi Fledel-Alon, Thomas L York, Nicholas R Polato, Kenneth M Olsen, Rasmus Nielsen, Susan R McCouch, Carlos D Bustamante, Michael D Purugganan
Affiliations
- PMID: 17907810
- PMCID: PMC1994709
- DOI: 10.1371/journal.pgen.0030163
Genome-wide patterns of nucleotide polymorphism in domesticated rice
Ana L Caicedo et al. PLoS Genet. 2007 Sep.
Abstract
Domesticated Asian rice (Oryza sativa) is one of the oldest domesticated crop species in the world, having fed more people than any other plant in human history. We report the patterns of DNA sequence variation in rice and its wild ancestor, O. rufipogon, across 111 randomly chosen gene fragments, and use these to infer the evolutionary dynamics that led to the origins of rice. There is a genome-wide excess of high-frequency derived single nucleotide polymorphisms (SNPs) in O. sativa varieties, a pattern that has not been reported for other crop species. We developed several alternative models to explain contemporary patterns of polymorphisms in rice, including a (i) selectively neutral population bottleneck model, (ii) bottleneck plus migration model, (iii) multiple selective sweeps model, and (iv) bottleneck plus selective sweeps model. We find that a simple bottleneck model, which has been the dominant demographic model for domesticated species, cannot explain the derived nucleotide polymorphism site frequency spectrum in rice. Instead, a bottleneck model that incorporates selective sweeps, or a more complex demographic model that includes subdivision and gene flow, are more plausible explanations for patterns of variation in domesticated rice varieties. If selective sweeps are indeed the explanation for the observed nucleotide data of domesticated rice, it suggests that strong selection can leave its imprint on genome-wide polymorphism patterns, contrary to expectations that selection results only in a local signature of variation.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Estimated Population Structure for 97 Accessions of O. sativa and O. rufipogon from 111 STS Loci
Vertical bars along the horizontal axis represent each Oryza accession; for all accessions, the proportion of ancestry under K = 7 clusters that can be attributed to each cluster is given by the length of each colored segment in a bar.
Figure 2. The Observed Marginal Derived Site-Frequency Spectra of Noncoding and Synonymous SNPs for Two Population Pairs: indica and O. rufipogon and tropical japonica and O. rufipogon
To accommodate SNPs with missing data, all spectra are plotted as the expected site frequency spectrum in a subsample of the data of size n = 16.
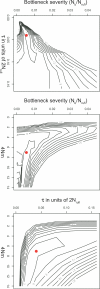
Figure 3. Contours of Composite Profile Log-Likelihood Surface under the Bottleneck and Migration (i.e., “Complex Demography”) Model for Three Key Demographic Parameters
Parameters include bottleneck severity, migration rate among demes (4_Nm_), and τ1 (time back until start of domestication scaled in units of 2_N_rufi). The maximum composite-likelihood estimate of the parameters is denoted by a red filled circle.
Figure 4. Observed and Expected Derived Site-Frequency Spectra under Various Models
The observed derived site-frequency spectrum for (A) indica and (B) tropical japonica, along with the expected site-frequency spectrum under the simple bottleneck, bottleneck plus migration demography, and bottleneck plus sweeps models. (C) Observed site-frequency spectrum for O. rufipogon and expected frequencies using a standard neutral model and a bottleneck plus migration model.
Figure 5. Composite Likelihood Surfaces in indica and tropical japonica under Models Incorporating Selection
A density plot of the marginal composite log-likelihood surface of the parameters α and κ, with the bottleneck severity υ fixed to its estimate, under the bottleneck plus sweeps model for (A) indica and (B) tropical japonica. The composite log-likelihood surface of the parameters α and κ under the pure selection model for (C) indica and (D) tropical japonica. The composite log-likelihood is represented as a deviation from the maximum log-likelihood, with lighter values representing higher composite likelihoods. Numbers above (A) and (B) indicate the total number of sweeps in the rice genome corresponding to each value of κ, and numbers to the right of (B) and (D) represent the selection coefficient, s, corresponding to each value of α, substituting an effective recombination rate of r = 10−12 and ln(2_N_) = 10 into the expression: α ≈ _rs_−1 ln(2N), then solving for s.
Similar articles
- Deleterious Variants in Asian Rice and the Potential Cost of Domestication.
Liu Q, Zhou Y, Morrell PL, Gaut BS. Liu Q, et al. Mol Biol Evol. 2017 Apr 1;34(4):908-924. doi: 10.1093/molbev/msw296. Mol Biol Evol. 2017. PMID: 28087781 - Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice.
Zhu Q, Zheng X, Luo J, Gaut BS, Ge S. Zhu Q, et al. Mol Biol Evol. 2007 Mar;24(3):875-88. doi: 10.1093/molbev/msm005. Epub 2007 Jan 11. Mol Biol Evol. 2007. PMID: 17218640 - A map of rice genome variation reveals the origin of cultivated rice.
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B. Huang X, et al. Nature. 2012 Oct 25;490(7421):497-501. doi: 10.1038/nature11532. Epub 2012 Oct 3. Nature. 2012. PMID: 23034647 Free PMC article. - New insights into the history of rice domestication.
Kovach MJ, Sweeney MT, McCouch SR. Kovach MJ, et al. Trends Genet. 2007 Nov;23(11):578-87. doi: 10.1016/j.tig.2007.08.012. Epub 2007 Oct 25. Trends Genet. 2007. PMID: 17963977 Review. - The complex history of the domestication of rice.
Sweeney M, McCouch S. Sweeney M, et al. Ann Bot. 2007 Nov;100(5):951-7. doi: 10.1093/aob/mcm128. Epub 2007 Jul 6. Ann Bot. 2007. PMID: 17617555 Free PMC article. Review.
Cited by
- Evolution of neutral and flowering genes along pearl millet (Pennisetum glaucum) domestication.
Lakis G, Navascués M, Rekima S, Simon M, Remigereau MS, Leveugle M, Takvorian N, Lamy F, Depaulis F, Robert T. Lakis G, et al. PLoS One. 2012;7(5):e36642. doi: 10.1371/journal.pone.0036642. Epub 2012 May 14. PLoS One. 2012. PMID: 22606277 Free PMC article. - Evolutionary Genomics of Structural Variation in Asian Rice (Oryza sativa) Domestication.
Kou Y, Liao Y, Toivainen T, Lv Y, Tian X, Emerson JJ, Gaut BS, Zhou Y. Kou Y, et al. Mol Biol Evol. 2020 Dec 16;37(12):3507-3524. doi: 10.1093/molbev/msaa185. Mol Biol Evol. 2020. PMID: 32681796 Free PMC article. - Genome-wide DNA polymorphisms in low Phosphate tolerant and sensitive rice genotypes.
Mehra P, Pandey BK, Giri J. Mehra P, et al. Sci Rep. 2015 Aug 17;5:13090. doi: 10.1038/srep13090. Sci Rep. 2015. PMID: 26278778 Free PMC article. - The Role of Standing Variation in the Evolution of Weedines Traits in South Asian Weedy Rice (Oryza spp.).
Huang Z, Kelly S, Matsuo R, Li LF, Li Y, Olsen KM, Jia Y, Caicedo AL. Huang Z, et al. G3 (Bethesda). 2018 Nov 6;8(11):3679-3690. doi: 10.1534/g3.118.200605. G3 (Bethesda). 2018. PMID: 30275171 Free PMC article. - TriMEDB: a database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae.
Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K. Mochida K, et al. BMC Plant Biol. 2008 Jun 30;8:72. doi: 10.1186/1471-2229-8-72. BMC Plant Biol. 2008. PMID: 18590523 Free PMC article.
References
- Hancock JF. Plant evolution and the origin of crop species. Cambridge (Massachusetts): CABI Publishing; 2004. 313
- Mannion AM. Domestication and the origins of agriculture: an appraisal. Prog Phys Geogr. 1999;23:37–56.
- Darwin C. On the origin of species. London: John Murray; 1859.
- Darwin C. The variation of animals and plants under domestication. London: John Murray; 1868.
- Armelagos GJ, Harper KN. Genomics at the origins of agriculture, part one. Evol Anthropol. 2005;14:68–77.