LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence - PubMed (original) (raw)
LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence
Kristie M Keeney et al. Mol Microbiol. 2007 Nov.
Abstract
The bacterial pathogen Listeria monocytogenes replicates within the cytosol of mammalian cells. Mechanisms by which the bacterium exploits the host cytosolic environment for essential nutrients are poorly defined. L. monocytogenes is a lipoate auxotroph and must scavenge this critical cofactor, using lipoate ligases to facilitate attachment of the lipoyl moiety to metabolic enzyme complexes. Although the L. monocytogenes genome encodes two putative lipoate ligases, LplA1 and LplA2, intracellular replication and virulence require only LplA1. Here we show that LplA1 enables utilization of host-derived lipoyl peptides by L. monocytogenes. LplA1 is dispensable for growth in the presence of free lipoate, but necessary for growth on low concentrations of mammalian lipoyl peptides. Furthermore, we demonstrate that the intracellular growth defect of the DeltalplA1 mutant is rescued by addition of exogenous lipoic acid to host cells, suggesting that L. monocytogenes dependence on LplA1 is dictated by limiting concentrations of available host lipoyl substrates. Thus, the ability of L. monocytogenes and other intracellular pathogens to efficiently use host lipoyl peptides as a source of lipoate may be a requisite adaptation for life within the mammalian cell.
Figures
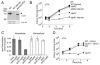
Fig. 1. L. monocytogenes has two functional lipoate ligase activities
A. Equivalent numbers of stationary phase wild type (WT), Δ_lplA1_, Δ_lplA2_ and lplA1::Tn917_Δ_lplA2 L. monocytogenes, based on OD600, were pelleted and protein harvested using the FastProtein™ Blue Matrix (MP Biomedicals). Bacterial lysates were analysed by SDS-PAGE, followed by immunoblot with an anti-lipoic acid antibody. The band slightly below 75 kDa has previously been identified by Mass-Spectrometry as lipoyl-E2-PDH (O'Riordan et al., 2003). Relative intensities of the bands to wild type were calculated using ImageJ. Loading of equivalent bacterial lysates was confirmed by stripping the blot and reprobing with an anti-E2 antibody (Stein and Firshein, 2000). Bacteria were grown in both the rich medium BHI and IMM, a medium that is limiting for lipoate; lplA1::Tn917_Δ_lplA2 was only studied in BHI as it exhibited impaired growth in IMM. B. Wild-type L. monocytogenes and the Δ_lplA1_ and Δ_lplA2_ mutant strains were grown in IMM containing different concentrations of lipoic acid. The OD600 was measured after bacteria had reached stationary phase (30 h) and plotted against lipoic acid concentration. The OD600 was measured in a Bioscreen Growth Curve Analyzer.
Fig. 2. Bacterial virulence in vivo requires LplA1, but not LplA2
A. 2 × 105 total cfu of exponentially growing cultures of wild-type and mutant L. monocytogenes were injected i.p. into ten 5- to 7-week-old male C57BL/6 mice. After 72 h, spleens and livers were harvested, homogenized and plated onto LB. B. Exponentially growing cultures of wild-type and mutant L. monocytogenes were mixed at a 1:10 (WT : Δ_lplA1_) or 1:1 ratio (WT : Δ_lplA2_) and 2 × 105 total cfu injected i.p. into six 5- to 7-week-old male C57BL/6 mice. After 72 h, spleens and livers were harvested, homogenized and plated onto LB with or without 1 μg ml−1 erm. The competitive index was calculated by dividing the number of wild-type strain cfu (ermS) by the number of mutant cfu (ermR). The horizontal line represents the median value. Statistically significant differences between two groups were determined by the Student's _t_-test at P < 0.05, as indicated by the symbol ‘*’.
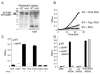
Fig. 3. LplA1, but not LplA2, is essential for intracellular growth
A. A culture of wild-type L. monocytogenes grown in IMM with or without lipoic acid was grown at 37°C to stationary phase, and protein was harvested as in Fig. 1. Bacterial lysates were analysed by immunoblot using an anti-lipoic acid antibody. The 75 kDa band (black arrowhead) corresponds to the L. monocytogenes E2-PDH. Porcine PDH was used as a positive control for the anti-lipoic acid antibody. Loading of equivalent bacterial lysates was confirmed by reprobing the blot with polyclonal anti-Listeria antibody. B. Wild-type, Δ_lplA1_ and Δ_lplA2_ bacterial strains were grown in IMM in the presence (dashed lines) or absence (solid lines) of lipoic acid overnight at 37°C, and used to infect J774 cell cultures. Intracellular growth was quantified by enumerating cfu. C. Wild-type bacteria were grown in BHI overnight at 37°C and used to infect J774 cells. At 6 h post infection, WT infected J774 cells were lysed and bacteria isolated. Quantitative RT-PCR was performed in triplicate on isolated bacterial RNA to determine template quantities of rpoA, lplA1 and lplA2. Template quantities were normalized against rpoA levels. To control for genomic DNA contamination, a portion of each RNA sample was removed from the reaction prior addition of reverse transcriptase (labelled ‘no RT’) and analysed by QRT-PCR. D. Starved bacterial strains were used to infect J774 cells with (dashed lines) or without (solid lines) 50 mM DHLA, then cells were lysed and cfu were enumerated. For all growth curves, the mean ± SD was calculated for each time point (n = 3).
Fig. 4. LplA1 enables utilization of degraded host-derived PDH for bacterial growth
A. J774 cell lysate was separated into cytosolic and mitochondrial fractions as described in Experimental procedures. Mitochondrial fractions and concentrated cytosolic fractions (100×) were analysed by immunoblot using a rabbit polyclonal anti-lipoic acid antibody. The 70 kDa band corresponds to mammalian E2-PDH, while the 55 kDa band corresponds to mammalian E2-KGDH. B. Wild-type (WT) L. monocytogenes was grown in IMM without lipoic acid supplemented with proteinase K (ProK)-digested, trypsin (Tryp)-digested or undigested porcine PDH. Growth was measured by OD600 and the mean value ± SD was calculated for each time point (n = 3). C. Wild-type and Δ_lplA1 L. monocytogenes_ were grown in IMM containing undigested or ProK-digested porcine PDH or KGDH at 5 mg l−1. After 35 h of growth in the Bioscreen Growth Curve Analyzer, bacterial cultures had reached stationary phase, and the OD600 values were plotted for each condition. BSA at 5 mg l−1 was also digested with ProK as a negative control. Mean values ± SD were calculated for each time point (n = 3). D. Wild-type, Δ_lplA1_ and Δ_lplA1_ complemented with a plasmid expressing LplA1 were grown in IMM containing the lipoate sources indicated; ProK-digested BSA at 5 mg l−1 was used as a negative control. Free lipoic acid (206 Da) (5 μg l−1) and ProK-digested porcine PDH (5 mg l−1) were dialysed against a 100 Da or a 500 Da MWCO membrane, and the retentate was used to supplement IMM. After 19.5 h of growth in conical tubes, the bacterial cultures had reached stationary phase; OD600 values for this time point were plotted for each condition. Growth was not determined (ND) for BSA and LA filtration experiments for Δ_lplA1_ and Δ_lplA1_ complemented with a plasmid expressing LplA1.
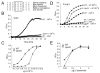
Fig. 5. LplA1 is required for optimal growth on small lipoyl peptides
A. The amino acid sequences of the dihydrolipoyl transacylase lipoyl domain from Homo sapiens (Accession No. AAA64512), dihydrolipoamide S-acetyltransferase lipoyl domain of Bos taurus (Accession No. XP_588501), dihydrolipoamide branched chain transacylase E2 lipoyl domain of Mus musculus (Accession No. NP_034152), dihydrolipoamide S-acetyltransferase lipoyl domain of Rattus norvegicus (Accession No. AAI07441) and the dihydrolipoamide acetyltransferase lipoyl domain of Sus scrofa (Accession No. NP_999159) aligned. B. Wild-type (WT) L. monocytogenes and the Δ_lplA1_ mutant strain were grown in IMM containing 5 μg l−1 DKL.A, or 5 μg l−1 non-lipoylated DKA. The OD600 was measured over time in a Bioscreen Growth Curve Analyzer and plotted as a function of time. C. Wild-type L. monocytogenes and the Δ_lplA1_ mutant strain were grown as in (B), but in IMM containing different concentrations of DKL.A. The OD600 was measured after bacteria had reached stationary phase (25 h) and plotted against lipoyl peptide concentration. D. Bacterial growth curves were performed as in (B), but 0.5 μg l−1 tripeptide (lipoylated or non-lipoylated) was added with or without prior aminopeptidase M digestion as indicated. E. Bacterial growth curves were performed as described in (B), but in IMM containing the concentrations of lipoamide indicated. After 30 h of growth, bacteria had reached stationary phase, and OD600 values were plotted against lipoamide concentration. The mean value ± SD was calculated for each time point (n = 3) in (B–E).
Fig. 6
Complementation of an E. coli strain deficient in lipoate utilization by L. monocytogenes (L.m.) LplA1. E.coli TM131 (lplA−lipA−) transformed with an empty IPTG-inducible vector, or the same vector expressing E. coli LplA or L. monocytogenes LplA1. TM131 is deficient in lipoate biosynthesis as well as endogenous LplA; growth requires either exogenous expression of a lipoate ligase, or supplementation with acetate and succinate. Clones expressing either empty vector, E.coli LplA or L. monocytogenes LplA1 were streaked on M9 minimal medium plates containing IPTG and free lipoic acid. Acetate and succinate were included in (A), but not (B) and (C). The boxes illustrated in (B) are magnified in (C) for viewing of single colonies. Some putative E. coli revertants were observed (large white colony observed in the insert of the LplA1-expressing E.coli strain).
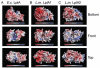
Fig. 7
Structural Modelling of L. monocytogenes (L.m.) LplA1 and LplA2. Electrostatic surface potentials for the crystallographic structure E. coli (E.c.) LplA (PDB ID 1X2H) and the modelled structures of LplA1 and LplA2 were calculated using APBS (Baker et al., 2001) and mapped onto their respective solvent accessible surfaces using Pymol (DeLano, 2002). Negative potentials (−10 kT e−1) are shown in red, positive potentials (10 kT e−1) in blue. The views for individual molecules are separated by a 90° rotation about the _x_-axis. The protein structures are shown at the same magnification and orientation for each view.
Similar articles
- Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein.
Morris TW, Reed KE, Cronan JE Jr. Morris TW, et al. J Bacteriol. 1995 Jan;177(1):1-10. doi: 10.1128/jb.177.1.1-10.1995. J Bacteriol. 1995. PMID: 8002607 Free PMC article. - Listeria intracellular growth and virulence require host-derived lipoic acid.
O'Riordan M, Moors MA, Portnoy DA. O'Riordan M, et al. Science. 2003 Oct 17;302(5644):462-4. doi: 10.1126/science.1088170. Science. 2003. PMID: 14564012 - Chlamydia trachomatis serovar L2 can utilize exogenous lipoic acid through the action of the lipoic acid ligase LplA1.
Ramaswamy AV, Maurelli AT. Ramaswamy AV, et al. J Bacteriol. 2010 Dec;192(23):6172-81. doi: 10.1128/JB.00717-10. Epub 2010 Sep 24. J Bacteriol. 2010. PMID: 20870766 Free PMC article. - ActA of Listeria monocytogenes and Its Manifold Activities as an Important Listerial Virulence Factor.
Pillich H, Puri M, Chakraborty T. Pillich H, et al. Curr Top Microbiol Immunol. 2017;399:113-132. doi: 10.1007/82_2016_30. Curr Top Microbiol Immunol. 2017. PMID: 27726006 Review. - From hot dogs to host cells: how the bacterial pathogen Listeria monocytogenes regulates virulence gene expression.
Freitag NE. Freitag NE. Future Microbiol. 2006 Jun;1(1):89-101. doi: 10.2217/17460913.1.1.89. Future Microbiol. 2006. PMID: 17661688 Review.
Cited by
- Staphylococcus aureus Tissue Infection During Sepsis Is Supported by Differential Use of Bacterial or Host-Derived Lipoic Acid.
Zorzoli A, Grayczyk JP, Alonzo F 3rd. Zorzoli A, et al. PLoS Pathog. 2016 Oct 4;12(10):e1005933. doi: 10.1371/journal.ppat.1005933. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27701474 Free PMC article. - Metabolism of the Gram-Positive Bacterial Pathogen Listeria monocytogenes.
Sauer JD, Herskovits AA, O'Riordan MXD. Sauer JD, et al. Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0066-2019. doi: 10.1128/microbiolspec.GPP3-0066-2019. Microbiol Spectr. 2019. PMID: 31418407 Free PMC article. Review. - Branched-chain fatty acids promote Listeria monocytogenes intracellular infection and virulence.
Sun Y, O'Riordan MX. Sun Y, et al. Infect Immun. 2010 Nov;78(11):4667-73. doi: 10.1128/IAI.00546-10. Epub 2010 Sep 7. Infect Immun. 2010. PMID: 20823206 Free PMC article. - Disruption of the adh (Acetoin Dehydrogenase) Operon Has Wide-Ranging Effects on Streptococcus mutans Growth and Stress Response.
Zuber P, Nakano MM, Kajfasz JK, Lemos JA. Zuber P, et al. J Bacteriol. 2022 Mar 15;204(3):e0057821. doi: 10.1128/jb.00578-21. Epub 2022 Jan 10. J Bacteriol. 2022. PMID: 35007154 Free PMC article. - Substrate Analogues Entering the Lipoic Acid Salvage Pathway via Lipoate-Protein Ligase 2 Interfere with Staphylococcus aureus Virulence.
Scattolini A, Grammatoglou K, Nikitjuka A, Jirgensons A, Mansilla MC, Windshügel B. Scattolini A, et al. ACS Infect Dis. 2024 Jun 14;10(6):2172-2182. doi: 10.1021/acsinfecdis.4c00148. Epub 2024 May 9. ACS Infect Dis. 2024. PMID: 38724014 Free PMC article.
References
- Akiba S, Matsugo S, Packer L, Konishi T. Assay of protein-bound lipoic acid in tissues by a new enzymatic method. Anal Biochem. 1998;258:299–304. - PubMed
- Baker H, Deangelis B, Baker ER, Hutner SH. A practical assay of lipoate in biologic fluids and liver in health and disease. Free Radic Biol Med. 1998;25:473–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI064540-01/AI/NIAID NIH HHS/United States
- R01 AI064540/AI/NIAID NIH HHS/United States
- R01 AI064540-02/AI/NIAID NIH HHS/United States
- T32AI007528/AI/NIAID NIH HHS/United States
- R01 AI064540-04/AI/NIAID NIH HHS/United States
- R01 AI064540-03/AI/NIAID NIH HHS/United States
- T32 AI007528/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases