Cro's role in the CI Cro bistable switch is critical for {lambda}'s transition from lysogeny to lytic development - PubMed (original) (raw)
Cro's role in the CI Cro bistable switch is critical for {lambda}'s transition from lysogeny to lytic development
Rachel A Schubert et al. Genes Dev. 2007.
Abstract
CI represses cro; Cro represses cI. This double negative feedback loop is the core of the classical CI-Cro epigenetic switch of bacteriophage lambda. Despite the classical status of this switch, the role in lambda development of Cro repression of the P(RM) promoter for CI has remained unclear. To address this, we created binding site mutations that strongly impaired Cro repression of P(RM) with only minimal effects on CI regulation of P(RM). These mutations had little impact on lambda development after infection but strongly inhibited the transition from lysogeny to the lytic pathway. We demonstrate that following inactivation of CI by ultraviolet treatment of lysogens, repression of P(RM) by Cro is needed to prevent synthesis of new CI that would otherwise significantly impede lytic development. Thus a bistable CI-Cro circuit reinforces the commitment to a developmental transition.
Figures
Figure 1.
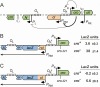
Cro repression of _P_RM. (A) Regulation of lytic and lysogenic promoters by Cro. (B) Repression of _P_RM by single-copy cro in cis. The chromosomally integrated _P_RM.lacZ transcriptional reporter carries the _O_R.cro region inserted so that lacZ is transcribed from _P_RM but is translated from the wild-type lacZ ribosome-binding site (RBS). Wild-type Cro, or a mutant protein carrying a deletion within its HTH DNA-binding motif (croΔ21), is expressed in cis from _P_R. (C) Repression of CI expression by single-copy cro in cis. The _P_RM.cI translational reporter is as in B, except that codon 20 of cI is fused to codon 10 of lacZ, such that lacZ is transcribed from _P_RM and translated from the cI RBS. Errors are 95% confidence intervals.
Figure 2.
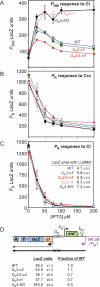
_O_R mutations relieving Cro repression of _P_RM. (A) The _P_RM-_O_R-P_R region and the mutations used in this study. The c21, r1, r314A, c12, and r204 mutations change the Δ_G of Cro binding to O_R1 by +1.7, +0.4, +1.7, +1.3, and +1.4 kcal/mol, respectively (Takeda et al. 1989), and the Δ_G of CI binding to _O_R1 by −0.2, +2.9, +1.0, −0.8, and +0.3 kcal/mol, respectively (Sarai and Takeda 1989). (B) Activity of the _P_RM. lacZ reporter (croΔ21 as in Fig. 1B) carrying KO mutations in _O_R3 and _O_R2, in response to Cro supplied from an IPTG-inducible expression plasmid (or an empty vector, dashed curve). LacZ activities have been normalized to aid comparison of repression with the _O_R2-KO mutation, which decreases _P_RM intrinsic activity by approximately twofold. (C) Effect of _O_R mutations on repression of _P_RM by Cro expressed from a single-copy cro gene in cis. The P_RM.lacZ reporters are as in Figure 1B, except that Δ_cro here indicates truncation of cro at the +43 position of _P_R. Fold repression is calculated without subtraction of background. (B,C) Errors are 95% confidence intervals.
Figure 3.
Effects of the mutations on other aspects of _O_R regulation. Response of _P_RM.lacZ reporters (A) and _P_R.lacZ reporters (B,C) to CI (A,C), or Cro (B) supplied by IPTG-inducible plasmids. The λ fragment extends from the +62 of _P_RM to the +43 of P_R. The inset in C shows reporter expression with CI supplied from a λ_att80 prophage. (D) Translational fusion reporters showing the effect of the _O_R mutations on cI translation from _P_RE mRNA. λ _P_RE is substituted by the constitutive 186 pB promoter. (A–D) Error bars and ranges are 95% confidence limits.
Figure 4.
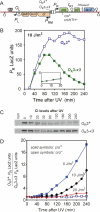
Behavior of the O_R mutant phages. (A) Phage production after infection. C600 cells were infected at a phage:bacterium ratio <0.01 with λ_WT or O_R mutant phages and assayed for IC (lysing cells + FP) at various times. Values are normalized to preburst averages. (B) CI Western blotting of extracts from C600 nonlysogenic cells (non) or single C600 lysogens of λ_WT or _O_R mutant phages. (Note that there is a cross-reacting host band running just below CI). Values are CI levels relative to wild type (with 95% confidence limits) obtained by quantitation of the blots (n = 8 or 9). (C) Phage production after UV irradiation (10 J/m2) of C600 lysogens. (D) Temperature induction of λ.cIts and λ.cIts._O_R3-x3 prophages. C600 lysogens were transferred from 30°C to 39°C and samples were assayed for FPs. (E) UV induction (as in C) of λ._O_R3-r1 and λ._O_R3-KO prophages and their P_RE_− derivatives. For the O_R3-r1 P_RE+ and _O_R3-r1 P_RE_− phages, respectively, 56% and 50% of lysogens were induced, producing 26 and 32 phage per irradiated lysogen. For the O_R3-KO P_RE+ and _O_R3-KO P_RE_− phages, respectively, 18% and 30% of lysogens were induced, producing on average 0.6 and 0.9 phage per irradiated lysogen.
Figure 5.
Effect of the _O_R3-x3 mutation on UV induction of the CI–Cro switch. (A) Structure of the chromosomally integrated cI._P_RM._P_R.cro.lacZ reporter constructs. LacZ activity from P_R (B), and Western blotting of CI (C) following 10 J/m2 UV treatment of the cro+ reporters. (D) Relative LacZ activities in the O_R3+ and _O_R3-x3 reporters after different UV doses. The strains carrying the croHTH− mutation showed that the _O_R3-x3 mutation made no difference to _P_R LacZ activity in the absence of Cro (open symbols, 10 J/m2 UV treatment).
Figure 6.
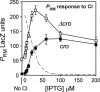
Repression of _P_RM by single-copy cro in cis in the presence of CI. The cro+ curve was obtained using the P_RM.lacZ transcriptional reporter in Figure 1B. The reporter for the Δ_cro curve is the same except that the cro gene is truncated at the +43 position of _P_R. CI was supplied with an IPTG-inducible expression plasmid system, with 150 μM IPTG producing a _P_RM response equivalent to that at lysogenic CI levels (Dodd et al. 2001). Errors are 95% confidence intervals. The dashed line shows the response of a P_R.lacZ (Δ_cro) reporter to CI (LacZ values divided by 5) (Dodd et al. 2004).
Similar articles
- Role of the lytic repressor in prophage induction of phage lambda as analyzed by a module-replacement approach.
Atsumi S, Little JW. Atsumi S, et al. Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4558-63. doi: 10.1073/pnas.0511117103. Epub 2006 Mar 14. Proc Natl Acad Sci U S A. 2006. PMID: 16537413 Free PMC article. - The Developmental Switch in Bacteriophage λ: A Critical Role of the Cro Protein.
Lee S, Lewis DEA, Adhya S. Lee S, et al. J Mol Biol. 2018 Jan 5;430(1):58-68. doi: 10.1016/j.jmb.2017.11.005. Epub 2017 Nov 20. J Mol Biol. 2018. PMID: 29158090 Free PMC article. - On the role of Cro in lambda prophage induction.
Svenningsen SL, Costantino N, Court DL, Adhya S. Svenningsen SL, et al. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4465-9. doi: 10.1073/pnas.0409839102. Epub 2005 Feb 23. Proc Natl Acad Sci U S A. 2005. PMID: 15728734 Free PMC article. - Research on phage λ: a lucky choice.
Lewis DEA, Adhya S. Lewis DEA, et al. EcoSal Plus. 2024 Dec 12;12(1):eesp00142023. doi: 10.1128/ecosalplus.esp-0014-2023. Epub 2023 Dec 22. EcoSal Plus. 2024. PMID: 39665539 Free PMC article. Review. - Switches in bacteriophage lambda development.
Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Oppenheim AB, et al. Annu Rev Genet. 2005;39:409-29. doi: 10.1146/annurev.genet.39.073003.113656. Annu Rev Genet. 2005. PMID: 16285866 Review.
Cited by
- Using axenic and gnotobiotic insects to examine the role of different microbes on the development and reproduction of the kissing bug Rhodnius prolixus (Hemiptera: Reduviidae).
Gilliland CA, Patel V, McCormick AC, Mackett BM, Vogel KJ. Gilliland CA, et al. Mol Ecol. 2023 Feb;32(4):920-935. doi: 10.1111/mec.16800. Epub 2022 Dec 27. Mol Ecol. 2023. PMID: 36464913 Free PMC article. - A λ Cro-Like Repressor Is Essential for the Induction of Conjugative Transfer of SXT/R391 Elements in Response to DNA Damage.
Poulin-Laprade D, Burrus V. Poulin-Laprade D, et al. J Bacteriol. 2015 Dec;197(24):3822-33. doi: 10.1128/JB.00638-15. Epub 2015 Oct 5. J Bacteriol. 2015. PMID: 26438816 Free PMC article. - Complete genome sequence of bacteriophage VvAW1, which infects Vibrio vulnificus.
Nigro OD, Culley AI, Steward GF. Nigro OD, et al. Stand Genomic Sci. 2012 Jul 30;6(3):415-26. doi: 10.4056/sigs.2846206. Epub 2012 Jul 20. Stand Genomic Sci. 2012. PMID: 23408718 Free PMC article. - Characterization of the virome of Paracoccus spp. (Alphaproteobacteria) by combined in silico and in vivo approaches.
Decewicz P, Dziewit L, Golec P, Kozlowska P, Bartosik D, Radlinska M. Decewicz P, et al. Sci Rep. 2019 May 27;9(1):7899. doi: 10.1038/s41598-019-44460-4. Sci Rep. 2019. PMID: 31133656 Free PMC article. - Development of an Anti-Acinetobacter baumannii Biofilm Phage Cocktail: Genomic Adaptation to the Host.
Blasco L, Bleriot I, González de Aledo M, Fernández-García L, Pacios O, Oliveira H, López M, Ortiz-Cartagena C, Fernández-Cuenca F, Pascual Á, Martínez-Martínez L, Pachón J, Azeredo J, Tomás M. Blasco L, et al. Antimicrob Agents Chemother. 2022 Mar 15;66(3):e0192321. doi: 10.1128/AAC.01923-21. Epub 2022 Jan 18. Antimicrob Agents Chemother. 2022. PMID: 35041503 Free PMC article.
References
- Adhya S., Gottesman M., Gottesman M. Promoter occlusion: Transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. - PubMed
- Baek K., Svenningsen S., Eisen H., Sneppen K., Brown S., Svenningsen S., Eisen H., Sneppen K., Brown S., Eisen H., Sneppen K., Brown S., Sneppen K., Brown S., Brown S. Single-cell analysis of λ immunity regulation. J. Mol. Biol. 2003;334:363–372. - PubMed
- Bailone A., Levine A., Devoret R., Levine A., Devoret R., Devoret R. Inactivation of prophage λ repressor in vivo. J. Mol. Biol. 1979;131:553–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases