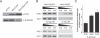MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae - PubMed (original) (raw)
MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae
Oliver Medvedik et al. PLoS Biol. 2007.
Abstract
Calorie restriction (CR) robustly extends the lifespan of numerous species. In the yeast Saccharomyces cerevisiae, CR has been proposed to extend lifespan by boosting the activity of sirtuin deacetylases, thereby suppressing the formation of toxic repetitive ribosomal DNA (rDNA) circles. An alternative theory is that CR works by suppressing the TOR (target of rapamycin) signaling pathway, which extends lifespan via mechanisms that are unknown but thought to be independent of sirtuins. Here we show that TOR inhibition extends lifespan by the same mechanism as CR: by increasing Sir2p activity and stabilizing the rDNA locus. Further, we show that rDNA stabilization and lifespan extension by both CR and TOR signaling is due to the relocalization of the transcription factors Msn2p and Msn4p from the cytoplasm to the nucleus, where they increase expression of the nicotinamidase gene PNC1. These findings suggest that TOR and sirtuins may be part of the same longevity pathway in higher organisms, and that they may promote genomic stability during aging.
Conflict of interest statement
Competing interests. DAS is a founder, director, share holder, and scientific consultant to Sirtris Pharmaceuticals, a company whose goal is to develop sirtuin-activating molecules to treat disease.
Figures
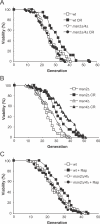
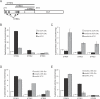
Figure 1. MSN2/4 Are Required for Lifespan Extension by CR and Rapamycin
(A) MSN2/4 are required for lifespan extension by CR (growth on 0.5% glucose). Average lifespans: W303 (wild-type [wt]) on 2% glucose, 25.0 divisions; W303 on CR diet, 30.8; _msn2_Δ/_4_Δ on 2% glucose, 24.2; and _msn2_Δ/_4_Δ on CR diet, 23.9. (B) The lifespans of _msn2_Δ and _msn4_Δ single mutants can be extended by CR. Average lifespans: _msn2_Δ on 2% glucose, 22.5 divisions; _msn2_Δ on CR diet, 29.9; _msn4_Δ on 2% glucose, 24.7; and _msn4_Δ on CR diet, 33.2. (C) MSN2/4 are required for lifespan extension by rapamycin (Rap). Average lifespans: wild-type, 23.3 divisions; wild-type + 1 nM rapamycin, 26.9; _msn2_Δ/_4_Δ, 23.8; and _msn2_Δ/_4_Δ + 1 nM rapamycin, 22.5.
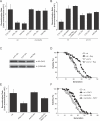
Figure 2. Effect of TOR Signaling on Sir2p Functions Requires MSN2/4 and PNC1
(A) Rapamycin and CR suppress rDNA recombination in an _MSN2/4_-dependent manner. (B) Deletion of PNC1 or treatment with NAM blocks the ability of CR or rapamycin treatment to lower rDNA recombination. (C) CR and rapamycin treatment do not increase Sir2p levels. (D) Deletion of PNC1 blocks the ability of rapamycin to extend lifespan. Average lifespans: wild-type (wt), 23.3 divisions; wild-type + 1 nM rapamycin, 26.9; _pnc1_Δ, 20.9; and _pnc1_Δ + 1 nM rapamycin, 21.4. (E) Overexpression (o/e) of PNC1 suppresses rDNA recombination in both wild-type and _msn2_Δ/_4_Δ strains. (F) Expression of PNC1 can extend the lifespan of a strain lacking MSN2/4. Average lifespans: wild-type, 23.2 divisions; wild-type + PNC1, 28.0; _msn2_Δ/_4_Δ, 22.0; and _msn2_Δ/_4_Δ + PNC1, 27.3. Glu, glucose; Rap, rapamycin.
Figure 3. MSN2/4 Regulate the Expression of PNC1
(A) Expression of Pnc1p-GFP is induced by CR and by rapamycin. (B) Western blotting shows that CR and rapamycin induce high levels of Pnc1p in an MSN2/4_-dependent manner. (C) A cdc25–10 mutant has increased expression of PNC1 but not if MSN2/4 are deleted. (D) Increased expression of PNC1 in response to salt stress, osmotic stress, and amino acid restriction requires MSN2/4, but increased expression of PNC1 in response to heat shock does not._ (E) MSN2/4 are not required for lifespan extension by heat shock. Average lifespans: wild-type at 30 °C, 22.2 divisions; wild-type at 37 °C, 26.8; _msn2_Δ/_4_Δ at 30 °C, 20.7; and _msn2_Δ/_4_Δ at 37 °C, 25.8. (F) Heat shock induction of PNC1 in an _msn2_Δ/_4_Δ strain is dependent upon HSF1. 2/4_Δ, msn2_Δ/_4_Δ; dox, doxycycline; Glu, glucose; low a.a., low amino acids; rap, rapamycin; Sorb, sorbitol; wt, wild-type.
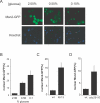
Figure 4. Msn2p Localizes to the Nucleus during CR
(A) CR promotes increased nuclear migration of Msn2p-GFP in a dose-dependent manner. (B) Quantification of the percent of cells with predominately nuclear Msn2p-GFP or Msn4p-GFP. (C) Deletion of TOR1 promotes nuclear localization of Msn2p-GFP. (D) A cdc25–10 mutant has increased nuclear localization of Msn2p-GFP.
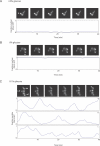
Figure 5. CR Induces Nucleo-Cytoplasmic Oscillations of Msn2p-GFP
During CR, Msn2p-GFP oscillates between the cytoplasm and nucleus. (A and B) Cells grown at 2% glucose concentrations (A) exhibit exclusively cytoplasmic localization of Msn2p-GFP. Cells placed in medium containing 0% glucose (B) exhibit exclusively nuclear localization of Msn2p-GFP. For cells in 2% glucose or 0% glucose, a plot of the ratio of average nuclear intensity versus average cytoplasmic intensity for a single representative cell, labeled with an arrowhead in the respective photomicrographs, is shown. (C) During CR conditions, cells exhibit nucleo-cytoplasmic oscillations of Msn2p-GFP with a periodicity of ∼2–3 min. A representative population of cells is shown at 0.1% glucose concentrations. The ratios of average nuclear intensity versus average cytoplasmic intensity for three representative cells, labeled a, b, and c, are shown plotted over the course of 20 min beneath the photomicrographs.
Figure 6. Deletion of MSN2/4 Abrogates Expression of LacZ Reporter Constructs Driven by the PNC1 Promoter
(A) Reporter constructs for analysis of STRE and Msn2p/4p function. (B) β-galactosidase activity of reporter constructs in wild-type (wt) cells grown in 2% YPD (Glu) (white bars), 0.5% YPD (black bars), or 2%YPD at 37 °C (grey bars). (C–E) Activity of reporter constructs in (C) an _msn2_Δ/_4_Δ strain, (D) an _msn2_Δ strain, or (E) an _msn4_Δ strain. Activity is listed as nanomoles of cleaved _o_-nitrophenol-β-D-galactopyranoside substrate per minute per milligram total protein.
Figure 7. Msn2p Directly Regulates PNC1
(A) Nuclear localization of Msn2p is sufficient to induce PNC1. (B) Chromatin immunoprecipitation analysis of Msn2p-HA binding to the PNC1 promoter. IN, input; IP, immunoprecipitation. (C) Relative enrichment of Msn2p-HA at the PNC1 promoter shows increased promoter binding during CR.
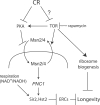
Figure 8. Model of How TOR Signaling and CR Regulate Yeast Replicative Lifespan
CR and TOR signaling regulate the nuclear localization of Msn2p/4p. When localized to the nucleus, Msn2p/4p promote the transcription of PNC1, a nicotinamidase. The removal of NAM by Pnc1p increases the activity of sirtuins, including Sir2p and Hst2p, which promote longevity by stabilizing the yeast rDNA array and preventing the formation of ERCs. TOR signaling may also regulate lifespan by sirtuin-independent pathways, such as the regulation of ribosomal biogenesis.
Similar articles
- Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins.
Smith DL Jr, McClure JM, Matecic M, Smith JS. Smith DL Jr, et al. Aging Cell. 2007 Oct;6(5):649-62. doi: 10.1111/j.1474-9726.2007.00326.x. Epub 2007 Aug 15. Aging Cell. 2007. PMID: 17711561 - Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae.
Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Anderson RM, et al. Nature. 2003 May 8;423(6936):181-5. doi: 10.1038/nature01578. Nature. 2003. PMID: 12736687 Free PMC article. - HST2 mediates SIR2-independent life-span extension by calorie restriction.
Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. Lamming DW, et al. Science. 2005 Sep 16;309(5742):1861-4. doi: 10.1126/science.1113611. Epub 2005 Jul 28. Science. 2005. PMID: 16051752 - The implication of Sir2 in replicative aging and senescence in Saccharomyces cerevisiae.
Ha CW, Huh WK. Ha CW, et al. Aging (Albany NY). 2011 Mar;3(3):319-24. doi: 10.18632/aging.100299. Aging (Albany NY). 2011. PMID: 21415463 Free PMC article. Review. - Angiotensin II blockade: how its molecular targets may signal to mitochondria and slow aging. Coincidences with calorie restriction and mTOR inhibition.
de Cavanagh EM, Inserra F, Ferder L. de Cavanagh EM, et al. Am J Physiol Heart Circ Physiol. 2015 Jul 1;309(1):H15-44. doi: 10.1152/ajpheart.00459.2014. Epub 2015 May 1. Am J Physiol Heart Circ Physiol. 2015. PMID: 25934099 Review.
Cited by
- Longevity and aging.
Kaeberlein M. Kaeberlein M. F1000Prime Rep. 2013;5:5. doi: 10.12703/P5-5. Epub 2013 Mar 4. F1000Prime Rep. 2013. PMID: 23513177 Free PMC article. - The role of feedback control mechanisms on the establishment of oscillatory regimes in the Ras/cAMP/PKA pathway in S. cerevisiae.
Besozzi D, Cazzaniga P, Pescini D, Mauri G, Colombo S, Martegani E. Besozzi D, et al. EURASIP J Bioinform Syst Biol. 2012 Jul 20;2012(1):10. doi: 10.1186/1687-4153-2012-10. EURASIP J Bioinform Syst Biol. 2012. PMID: 22818197 Free PMC article. - Hot topics in aging research: protein translation, 2009.
Kennedy BK, Kaeberlein M. Kennedy BK, et al. Aging Cell. 2009 Dec;8(6):617-23. doi: 10.1111/j.1474-9726.2009.00522.x. Epub 2009 Sep 11. Aging Cell. 2009. PMID: 19747234 Free PMC article. Review. - Could glucose be a proaging factor?
Kassi E, Papavassiliou AG. Kassi E, et al. J Cell Mol Med. 2008 Aug;12(4):1194-8. doi: 10.1111/j.1582-4934.2008.00329.x. Epub 2008 Apr 8. J Cell Mol Med. 2008. PMID: 18400054 Free PMC article. Review. - Role of TOR signaling in aging and related biological processes in Drosophila melanogaster.
Katewa SD, Kapahi P. Katewa SD, et al. Exp Gerontol. 2011 May;46(5):382-90. doi: 10.1016/j.exger.2010.11.036. Epub 2010 Dec 2. Exp Gerontol. 2011. PMID: 21130151 Free PMC article. Review.
References
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. - PubMed
- Barton A. Some aspects of cell division in Saccharomyces cerevisiae . J Gen Microbiol. 1950;4:84–86. - PubMed
- Sinclair DA, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. - PubMed
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. - PubMed
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AG19972/AG/NIA NIH HHS/United States
- R01 GM068072/GM/NIGMS NIH HHS/United States
- R01 AG028730/AG/NIA NIH HHS/United States
- R01 AG019972/AG/NIA NIH HHS/United States
- R01 AG019719-07/AG/NIA NIH HHS/United States
- R01 AG028730-02/AG/NIA NIH HHS/United States
- R01 AG028730-01A1/AG/NIA NIH HHS/United States
- R01GM068072/GM/NIGMS NIH HHS/United States
- R01 AG019719/AG/NIA NIH HHS/United States
- R01 AG019719-06A1/AG/NIA NIH HHS/United States
- R01 AG028730-03/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases