Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1 - PubMed (original) (raw)
Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1
Qiang Liu et al. Neuron. 2007.
Abstract
Mutations in the amyloid precursor protein (APP) cause early-onset Alzheimer's disease (AD), but the only genetic risk factor for late-onset AD is the varepsilon4 allele of apolipoprotein E (apoE), a major cholesterol carrier. Using Cre-lox conditional knockout mice, we demonstrate that lipoprotein receptor LRP1 expression regulates apoE and cholesterol levels within the CNS. We also found that deletion of APP and its homolog APLP2, or components of the gamma-secretase complex, significantly enhanced the expression and function of LRP1, which was reversed by forced expression of the APP intracellular domain (AICD). We further show that AICD, together with Fe65 and Tip60, interacts with the LRP1 promoter and suppresses its transcription. Together, our findings support that the gamma-secretase cleavage of APP plays a central role in regulating apoE and cholesterol metabolism in the CNS via LRP1 and establish a biological linkage between APP and apoE, the two major genetic determinants of AD.
Figures
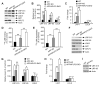
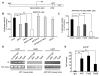
Figure 1. APP Regulates ApoE and Cholesterol Metabolism
(A and B) ApoE levels were decreased and cholesterol content was increased in the absence of APP or APP/APLP2. ApoE and cholesterol levels were measured in triplicates in lysates of WT, APP-KO and APP/APLP2-DKO MEF cells, normalized against total protein and plotted as a percentage of WT controls. (C and D) ApoE levels were also increased and cholesterol content decreased in brain lysates of APP-KO and APP/APLP2-DKO mice. ApoE and cholesterol levels were measured in brain lysates of WT, APP-KO and APP/APLP2-DKO mice (n=4), normalized against total protein and plotted as a percentage of WT controls. For this and subsequent figures, data represent mean ± SEM; N.S., not significant; *, P<0.05; **, P<0.01.
Figure 2. APP/APLP2 Regulates LRP1 Expression and Function
(A) LRP1 and LDLR expression levels were compared between WT, APP-KO and APP/APLP2-DKO MEF cells by Western blot. Equal amount of protein in this and subsequent figures was loaded to each lane. (B) Densitometric analyses of Western blots from triplicate samples demonstrate a significant increase in the expression of LRP1, but not LDLR, in the absence of APP/APLP2. (C) LRP1 and LDLR mRNA levels were quantified in WT, APP-KO and APP/APLP2-DKO MEF cells by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of APP/APLP2. (D) 125I-α2M (1 nM) binding to WT, APP-KO and APP/APLP2-DKO MEF cells was performed at 4°C for 1 h in the absence or presence of RAP (500 nM). RAP-inhibitable 125I-α2M binding was normalized against total cellular protein and plotted as a percentage of WT controls. (E) 125I-α2M (1 nM) uptake and degradation assays were performed at 37°C for 4 h in the absence or presence of RAP (500 nM). RAP-inhibitable 125I-α2M degradation was normalized against total cellular protein and plotted as a percentage of WT controls. (F) LRP1 and LDLR expression levels in the brain were compared between WT, APP-KO and APP/APLP2-DKO newborn mice by Western blot. (G) Densitometric analyses of Western blots (n=3) indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of APP or APP/APLP2. (H) LRP1 and LDLR mRNA levels were quantified in the brain of WT, APP-KO and APP/APLP2-DKO newborn mice (n=3) by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of APP/APLP2. (I) LRP1 expression in the brain was compared by Western blot between WT and APP-KO mice at 4 months of age. Similar increase in LRP1 expression was observed in adult APP-KO mouse brain.
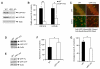
Figure 3. LRP1 Is Essential for Brain ApoE and Cholesterol Metabolism
(A) LRP1 expression in the forebrain was evaluated in LRP1 forebrain knockout (LRP1-KO) and WT littermate control mice by Western blot using two distinct LRP1 antibodies (against 515-kDa subunit and 85-kDa subunit, respectively). Equal amount of protein in this and subsequent figures was loaded to each lane. (B) Densitometric quantification of LRP1 expression was performed as described in Materials and Methods from two independent experiments (n=4). (C) Double immunofluorescence staining using an LRP1 antibody (detected with Alexa 488, green) and neuronal marker NeuN antibody (detected with Alexa 633, red). Shown are representative staining in CA1 neurons and pyramidal neurons of frontal cortex. Note that LRP1 expression was almost absent in these forebrain neurons. (D) APP-FL and APP-CTF levels were compared by Western blot between WT and LRP1-KO MEF cells. (E) APP-FL and APP-CTF levels were compared by Western blot between WT and LRP1-KO mouse brains. (F and G) ApoE and cholesterol levels in the brain were compared between LRP1-KO and WT controls (n=4). ApoE levels were significantly higher (F) and cholesterol levels were significantly lower (G) in LRP1-KO mice.
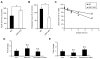
Figure 4. Absence of LRP1 Expression Increases ApoE Half-life
(A and B) ApoE (A) and cholesterol (B) levels were measured in triplicates in lysates of WT and LRP1-KO MEF cells, normalized against total protein and plotted as a percentage of WT controls. (C) WT and LRP1-KO MEF cells were incubated with serum-free medium for 2 h and chased in the presence of protein synthesis inhibitor cycloheximide for 0, 1, 2, or 4 h. ApoE levels under each condition were measured by ELISA and plotted against chase time. Note apoE half-life is significantly increased in LRP1-KO MEF cells. (D and E) ApoE mRNA was quantified by real-time PCR in WT, APP-KO, APP/APLP2-DKO MEF cells (D) or brain tissues (E). Note apoE mRNA levels were not significantly affected by APP/APLP2 deletion.
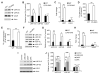
Figure 5. γ-Secretase Regulates LRP1 Expression
(A) LRP1 and LDLR expression levels were compared between WT and PS-DKO MEF cells by Western blot. (B) Densitometric analyses of Western blots from quadruplicate samples indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of PS. (C) LRP1 and LDLR mRNA levels were quantified in WT and PS-DKO MEF cells by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of PS. (D and E) 125I-α2M binding (D) and degradation assays (E) were performed in WT and PS-DKO MEF cells as described in Figure 2. (F) LRP1 and LDLR expression levels were also analyzed in WT and PS-DKO mouse brains by Western blot. Note the significant accumulation of APP CTF in PS-DKO mouse brains. (G) Densitometric analyses (n=3) indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of PS. (H) LRP1 and LDLR mRNA levels were quantified in WT and PS-DKO mouse brain (n=3) by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of PS. (I) WT MEF cells were treated with vehicle control or γ-secretase inhibitors DAPT (2 μM), DFK (100 μM), or L685,458 (1 μM) for 48 h. Expression levels of LRP1 and LDLR were measured by Western blot. (J) Densitometric analyses of Western blots from triplicate samples indicate an increase in the expression of LRP1, but not LDLR, upon γ-secretase inhibitor treatments.
Figure 6. AICD Rescues LRP1 Expression in the Absence of APP/APLP2 or PS1/2
(A) U87 cells were transiently transfected with plasmid constructs as indicated. LRP1 and LDLR expression levels were analyzed by Western blot. (B) Densitometric analyses of Western blots from triplicate samples indicate that a forced expression of AICD and Fe65 suppressed the expression of LRP1, but not LDLR. (C) PS-DKO MEF cells were transiently infected with retroviral vector control or AICD C50, C57, or C59 cDNA. LRP1 and LDLR expression levels were then analyzed by Western blot and compared with those of WT MEF cells. (D) Densitometric analyses of Western blot from triplicate samples indicate that AICD expression suppressed LRP1 expression close to the levels seen in WT MEF cells. (E) The rescuing effect of AICD on LRP1 expression was also observed at the mRNA levels as measured by real-time PCR. (F-H) Similar experiments to those shown in (C-E) were performed with APP/APLP2-DKO MEF cells.
Figure 7. AICD Binds To and Suppresses LRP1 Promoter Activation
(A) Schematic diagram of LRP1 promoter-luciferase construct. (B) BHK570 cells were transiently co-transfected with the LRP1 promoter-Luc construct together with control vector, AICD, Fe65, AICD/Fe65, AICD mutant (Y682G), AICD mutant/Fe65, or NICD. LRP1 promoter-driven luciferase activity was significantly decreased by AICD and Fe65 and further by the co-expression of both, but not by an AICD mutant or NICD. (C) APP/APLP2-DKO MEF cells were transiently co-transfected with the LRP1 promoter-Luc construct together with control vector, AICD, Fe65, or both, and the luciferase activity was measured as in (B). (D) ChIP assay showed that antibodies to AICD, Fe65 and Tip60, but not control IgG, immunoprecipitate LRP1 promoter DNA fragment. Notch target HES1 promoter was used as a negative control. The ability of anti-Fe65 and anti-Tip60 to immunoprecipitate LRP1 promoter DNA is greatly reduced in APP-KO mouse brain. (E) Quantitative real-time PCR analysis of LRP1 promoter DNA immunoprecipitated by control IgG or antibodies to AICD, Fe65 and Tip60.
Figure 8. AICD Rescues ApoE and Cholesterol Defects in Cells Deficient in PS or APP
(A-D) ApoE (A and B) and cholesterol levels (C and D) were measured in WT, PS-DKO and BACE1-KO MEF cells (A and C) and mouse brains (B and D). ApoE levels were decreased while cholesterol levels were increased in PS-DKO but not BACE1-KO MEF cells and mouse brain when compared to their WT controls. (E-H) APP/APLP2-DKO (E and G) and PS-DKO (F and H) MEF cells were transiently infected with retroviral vector control or AICD cDNA. LRP1 expression levels were then analyzed by Western blot, quantified by densitometry, and compared with those of WT MEF cells. Expression of AICD partially rescued apoE and cholesterol levels in both APP/APLP2-DKO and PSDKO MEF cells.
Similar articles
- Functional role of lipoprotein receptors in Alzheimer's disease.
Jaeger S, Pietrzik CU. Jaeger S, et al. Curr Alzheimer Res. 2008 Feb;5(1):15-25. doi: 10.2174/156720508783884675. Curr Alzheimer Res. 2008. PMID: 18288927 Review. - Blood-brain barrier-associated pericytes internalize and clear aggregated amyloid-β42 by LRP1-dependent apolipoprotein E isoform-specific mechanism.
Ma Q, Zhao Z, Sagare AP, Wu Y, Wang M, Owens NC, Verghese PB, Herz J, Holtzman DM, Zlokovic BV. Ma Q, et al. Mol Neurodegener. 2018 Oct 19;13(1):57. doi: 10.1186/s13024-018-0286-0. Mol Neurodegener. 2018. PMID: 30340601 Free PMC article. - The role of low-density receptor-related protein 1 (LRP1) as a competitive substrate of the amyloid precursor protein (APP) for BACE1.
von Einem B, Schwanzar D, Rehn F, Beyer AS, Weber P, Wagner M, Schneckenburger H, von Arnim CA. von Einem B, et al. Exp Neurol. 2010 Sep;225(1):85-93. doi: 10.1016/j.expneurol.2010.05.017. Epub 2010 May 26. Exp Neurol. 2010. PMID: 20685197 - Co-localization of the amyloid precursor protein and Notch intracellular domains in nuclear transcription factories.
Konietzko U, Goodger ZV, Meyer M, Kohli BM, Bosset J, Lahiri DK, Nitsch RM. Konietzko U, et al. Neurobiol Aging. 2010 Jan;31(1):58-73. doi: 10.1016/j.neurobiolaging.2008.03.001. Epub 2008 Apr 10. Neurobiol Aging. 2010. PMID: 18403052 Free PMC article. - γ-Secretase, apolipoprotein E and cellular cholesterol metabolism.
Walter J. Walter J. Curr Alzheimer Res. 2012 Feb;9(2):189-99. doi: 10.2174/156720512799361583. Curr Alzheimer Res. 2012. PMID: 21605030 Review.
Cited by
- Aspirin-triggered lipoxin A4 stimulates alternative activation of microglia and reduces Alzheimer disease-like pathology in mice.
Medeiros R, Kitazawa M, Passos GF, Baglietto-Vargas D, Cheng D, Cribbs DH, LaFerla FM. Medeiros R, et al. Am J Pathol. 2013 May;182(5):1780-9. doi: 10.1016/j.ajpath.2013.01.051. Epub 2013 Mar 16. Am J Pathol. 2013. PMID: 23506847 Free PMC article. - Peroxisome proliferator-activated receptors and Alzheimer's disease: hitting the blood-brain barrier.
Zolezzi JM, Inestrosa NC. Zolezzi JM, et al. Mol Neurobiol. 2013 Dec;48(3):438-51. doi: 10.1007/s12035-013-8435-5. Epub 2013 Mar 14. Mol Neurobiol. 2013. PMID: 23494748 Review. - Amyloid-beta Alzheimer targets - protein processing, lipid rafts, and amyloid-beta pores.
Arbor SC, LaFontaine M, Cumbay M. Arbor SC, et al. Yale J Biol Med. 2016 Mar 24;89(1):5-21. eCollection 2016 Mar. Yale J Biol Med. 2016. PMID: 27505013 Free PMC article. Review. - The role of apolipoprotein E in Alzheimer's disease.
Kim J, Basak JM, Holtzman DM. Kim J, et al. Neuron. 2009 Aug 13;63(3):287-303. doi: 10.1016/j.neuron.2009.06.026. Neuron. 2009. PMID: 19679070 Free PMC article. Review. - Lipoprotein receptors--an evolutionarily ancient multifunctional receptor family.
Dieckmann M, Dietrich MF, Herz J. Dieckmann M, et al. Biol Chem. 2010 Nov;391(11):1341-63. doi: 10.1515/BC.2010.129. Biol Chem. 2010. PMID: 20868222 Free PMC article. Review.
References
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG030128/AG/NIA NIH HHS/United States
- R01 AG027924/AG/NIA NIH HHS/United States
- R01 AG027924-02/AG/NIA NIH HHS/United States
- R37 HL063762/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous